Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
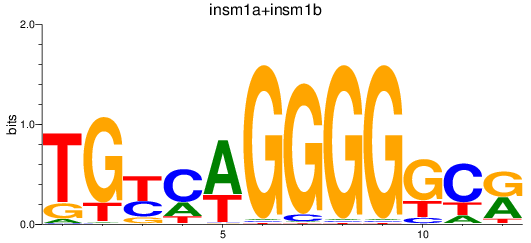
Results for insm1a+insm1b
Z-value: 0.62
Transcription factors associated with insm1a+insm1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
insm1b
|
ENSDARG00000053301 | insulinoma-associated 1b |
|
insm1a
|
ENSDARG00000091756 | insulinoma-associated 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| insm1b | dr11_v1_chr17_+_41463942_41463942 | 0.58 | 3.0e-01 | Click! |
| insm1a | dr11_v1_chr20_-_48485354_48485354 | 0.23 | 7.1e-01 | Click! |
Activity profile of insm1a+insm1b motif
Sorted Z-values of insm1a+insm1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_49566364 | 0.72 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr2_+_58221163 | 0.67 |
ENSDART00000157939
|
FO704813.1
|
|
| chr5_-_51830997 | 0.64 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr7_+_73397283 | 0.61 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr9_+_4429593 | 0.60 |
ENSDART00000184855
|
FP015810.1
|
|
| chr17_-_4318393 | 0.59 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr8_-_37391372 | 0.47 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr7_+_23515966 | 0.44 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr4_+_13733838 | 0.44 |
ENSDART00000067166
ENSDART00000133157 |
cntn1b
|
contactin 1b |
| chr8_+_1082100 | 0.44 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr18_+_18612388 | 0.43 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr18_+_5137241 | 0.43 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr18_+_642889 | 0.41 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr15_-_33925851 | 0.41 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr10_+_21801030 | 0.40 |
ENSDART00000160881
ENSDART00000187969 |
pcdh1g30
|
protocadherin 1 gamma 30 |
| chr13_+_23176330 | 0.39 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr17_-_52579709 | 0.36 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr7_+_73649686 | 0.35 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr1_+_12767318 | 0.35 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr15_-_19128705 | 0.34 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr18_+_50278858 | 0.34 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr12_-_48992527 | 0.34 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr9_+_53719230 | 0.32 |
ENSDART00000165827
|
PCDH8
|
si:ch211-199f5.1 |
| chr12_+_11352630 | 0.31 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr23_+_19594608 | 0.29 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr7_-_41489997 | 0.29 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr21_+_32332616 | 0.29 |
ENSDART00000154105
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr3_-_6768905 | 0.29 |
ENSDART00000193638
ENSDART00000123809 ENSDART00000189440 ENSDART00000188335 |
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr6_+_3166941 | 0.27 |
ENSDART00000168912
|
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr13_+_40034176 | 0.27 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr16_-_29458806 | 0.27 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr24_-_28306484 | 0.26 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr20_+_1996202 | 0.25 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr17_+_45454943 | 0.25 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr23_-_21797517 | 0.25 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr14_+_8504410 | 0.24 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr5_+_57641554 | 0.24 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr11_+_24620742 | 0.24 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr17_+_6793001 | 0.23 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr10_-_41157135 | 0.23 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr21_+_30673183 | 0.22 |
ENSDART00000144652
|
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr19_+_2726819 | 0.22 |
ENSDART00000187122
ENSDART00000112414 |
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr9_+_29040425 | 0.22 |
ENSDART00000150201
|
MRAS
|
si:ch73-116o1.2 |
| chr4_-_25908871 | 0.22 |
ENSDART00000066949
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr22_-_881080 | 0.20 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr3_+_17346502 | 0.20 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr14_+_42172677 | 0.20 |
ENSDART00000148544
|
pcdh18b
|
protocadherin 18b |
| chr18_+_45573416 | 0.20 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr4_+_90048 | 0.20 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr1_-_40341306 | 0.19 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr18_+_48608366 | 0.19 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr23_+_16430559 | 0.19 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr25_-_21085661 | 0.19 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr1_-_21599219 | 0.18 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr18_+_45573251 | 0.18 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr6_-_10828880 | 0.18 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr4_-_5597802 | 0.18 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr9_-_4598883 | 0.17 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr7_-_72261721 | 0.17 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr4_+_7888047 | 0.16 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr12_+_30368145 | 0.16 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr10_+_41549819 | 0.15 |
ENSDART00000114210
|
CABZ01049847.1
|
|
| chr3_+_22079219 | 0.15 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr7_-_72423666 | 0.15 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr11_-_30352333 | 0.15 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr19_-_19556778 | 0.15 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr8_-_49705058 | 0.15 |
ENSDART00000083760
|
frmd3
|
FERM domain containing 3 |
| chr21_+_25068215 | 0.14 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr14_-_6402769 | 0.14 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr8_+_41453691 | 0.14 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr14_-_30876299 | 0.13 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr25_-_28443607 | 0.13 |
ENSDART00000157243
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr1_+_2112726 | 0.13 |
ENSDART00000131714
ENSDART00000138396 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr2_-_16216568 | 0.13 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr25_-_24046870 | 0.13 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr19_+_1097393 | 0.13 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr18_-_41211980 | 0.13 |
ENSDART00000171431
|
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr8_+_23521974 | 0.13 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr13_-_49666615 | 0.13 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr17_+_9310259 | 0.13 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr10_+_6383270 | 0.12 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr3_-_40051425 | 0.12 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr25_-_13659249 | 0.12 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr10_+_22527715 | 0.12 |
ENSDART00000134864
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr9_-_1434484 | 0.12 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr15_-_3078600 | 0.12 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr14_-_4076480 | 0.12 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr21_+_11105007 | 0.12 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr19_+_46158078 | 0.11 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_50371951 | 0.11 |
ENSDART00000184488
ENSDART00000171295 |
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr9_+_38427572 | 0.11 |
ENSDART00000108860
|
CYP27A1 (1 of many)
|
zgc:136333 |
| chr6_-_55354004 | 0.11 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr13_-_24874950 | 0.11 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr24_-_33873451 | 0.10 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr2_+_1202347 | 0.10 |
ENSDART00000075837
|
CABZ01084566.1
|
|
| chr3_+_18437397 | 0.10 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr19_-_33996277 | 0.10 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr21_+_21791799 | 0.10 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr7_-_30605089 | 0.09 |
ENSDART00000173775
ENSDART00000173789 ENSDART00000166046 ENSDART00000111626 |
rnf111
|
ring finger protein 111 |
| chr10_+_44940693 | 0.09 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr2_+_7132292 | 0.09 |
ENSDART00000153404
ENSDART00000012119 |
zgc:110366
|
zgc:110366 |
| chr24_-_2917540 | 0.09 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr3_-_43650189 | 0.08 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr12_+_47907277 | 0.08 |
ENSDART00000097714
|
tbata
|
thymus, brain and testes associated |
| chr3_-_43646733 | 0.08 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr8_-_1264893 | 0.08 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr12_+_47280839 | 0.08 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr15_+_1796313 | 0.07 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr21_-_35534401 | 0.07 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr25_+_16601839 | 0.07 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr3_+_20001608 | 0.07 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr21_-_30415524 | 0.07 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr24_-_10394277 | 0.07 |
ENSDART00000127568
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr13_+_30951155 | 0.06 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr11_+_19060278 | 0.06 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr11_-_23697217 | 0.06 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_+_15100742 | 0.06 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr21_+_5800306 | 0.06 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr3_-_15668433 | 0.05 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr9_+_41690153 | 0.05 |
ENSDART00000100226
|
ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr7_+_31871830 | 0.05 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr19_+_9032073 | 0.05 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr25_-_25575241 | 0.05 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr18_+_17493859 | 0.05 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr10_-_105100 | 0.05 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr18_-_127873 | 0.05 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr4_+_70791887 | 0.04 |
ENSDART00000164234
|
si:ch211-127b6.2
|
si:ch211-127b6.2 |
| chr15_-_47822597 | 0.04 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr17_+_25197180 | 0.04 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr11_-_26040594 | 0.04 |
ENSDART00000144115
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr14_-_30983011 | 0.04 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr12_-_4683325 | 0.04 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr2_+_21486529 | 0.04 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr22_+_17536989 | 0.03 |
ENSDART00000149531
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr21_-_43117327 | 0.03 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr3_-_61592417 | 0.03 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr1_-_18803919 | 0.03 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr13_-_2981472 | 0.03 |
ENSDART00000184820
|
CABZ01087623.1
|
|
| chr3_-_7129057 | 0.03 |
ENSDART00000125947
|
BX005085.1
|
|
| chr5_-_10236599 | 0.03 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr13_+_36680564 | 0.03 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr9_-_42989297 | 0.03 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr14_-_49992709 | 0.02 |
ENSDART00000159988
|
fam193b
|
family with sequence similarity 193, member B |
| chr20_+_1834262 | 0.02 |
ENSDART00000185316
|
CABZ01065423.1
|
|
| chr14_+_8174828 | 0.02 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr6_+_9421279 | 0.02 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr8_-_14484599 | 0.01 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr18_+_18000695 | 0.01 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr13_-_36680531 | 0.01 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr6_-_39051319 | 0.01 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr8_+_15251448 | 0.01 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr4_+_69577362 | 0.00 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr25_+_32496723 | 0.00 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr4_+_40329524 | 0.00 |
ENSDART00000165722
|
znf993
|
zinc finger protein 993 |
| chr25_-_25575717 | 0.00 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr17_+_43013171 | 0.00 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of insm1a+insm1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.4 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |