Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
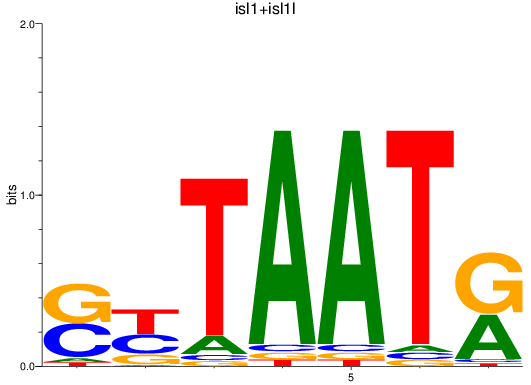
Results for isl1+isl1l
Z-value: 0.86
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
|
isl1l
|
ENSDARG00000110204 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1l | dr11_v1_chr10_+_8680730_8680730 | 0.10 | 8.7e-01 | Click! |
| isl1 | dr11_v1_chr5_-_40734045_40734045 | -0.01 | 9.9e-01 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_11457500 | 0.47 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr1_-_25679339 | 0.45 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr14_+_21222287 | 0.41 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr16_-_29387215 | 0.36 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr6_-_27123327 | 0.36 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr22_+_19220459 | 0.34 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr19_+_10855158 | 0.31 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr13_+_46941930 | 0.29 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr7_-_38664947 | 0.26 |
ENSDART00000100546
ENSDART00000112405 |
c6ast1
|
six-cysteine containing astacin protease 1 |
| chr6_-_346084 | 0.26 |
ENSDART00000162599
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr7_+_36035432 | 0.26 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr7_+_13988075 | 0.26 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr19_-_47570672 | 0.24 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr4_+_2230701 | 0.23 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr14_-_1454045 | 0.23 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr17_+_33375469 | 0.22 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr12_+_22672323 | 0.22 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr1_-_59348118 | 0.22 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr12_-_13205854 | 0.21 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr1_-_45146834 | 0.21 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr5_+_32345187 | 0.21 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr10_-_2943474 | 0.21 |
ENSDART00000188698
|
oclna
|
occludin a |
| chr5_-_41831646 | 0.20 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr11_-_438294 | 0.20 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr20_-_45709990 | 0.19 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr15_-_44052927 | 0.19 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr20_+_23440632 | 0.19 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr19_+_10331325 | 0.18 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr3_-_32956808 | 0.18 |
ENSDART00000183902
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr16_-_55259199 | 0.18 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr4_-_5826320 | 0.17 |
ENSDART00000165354
|
foxm1
|
forkhead box M1 |
| chr23_+_26142613 | 0.16 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr24_-_32665283 | 0.16 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr9_-_35633827 | 0.16 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr5_-_30615901 | 0.16 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr5_-_64203101 | 0.16 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr22_-_36875264 | 0.16 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr5_-_41838354 | 0.16 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr20_+_34770197 | 0.16 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr21_+_31253048 | 0.15 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr23_-_45398622 | 0.15 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr8_+_21254192 | 0.15 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr14_-_36799280 | 0.14 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr8_-_14121634 | 0.14 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr12_-_19028046 | 0.14 |
ENSDART00000142253
ENSDART00000078484 |
rangap1b
|
Ran GTPase activating protein 1b |
| chr8_+_53064920 | 0.14 |
ENSDART00000164823
|
nadka
|
NAD kinase a |
| chr7_+_38897836 | 0.14 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr3_-_56871984 | 0.14 |
ENSDART00000189908
ENSDART00000183146 ENSDART00000183087 |
zgc:112148
|
zgc:112148 |
| chr17_-_4252221 | 0.14 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr5_+_31168096 | 0.14 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr3_+_15505275 | 0.13 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr7_+_4538334 | 0.13 |
ENSDART00000139326
ENSDART00000146915 |
si:dkey-83f18.9
|
si:dkey-83f18.9 |
| chr19_-_6193448 | 0.13 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr17_+_39741926 | 0.13 |
ENSDART00000154996
ENSDART00000154599 |
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr20_-_35012093 | 0.13 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr20_-_25522911 | 0.13 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr20_+_2281933 | 0.13 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr14_+_1240419 | 0.12 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr17_-_2036850 | 0.12 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr14_+_16287968 | 0.12 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr20_+_37794633 | 0.12 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr4_-_75707991 | 0.12 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr7_+_53541173 | 0.12 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr16_+_52999778 | 0.12 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr12_-_13205572 | 0.11 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr4_+_17280868 | 0.11 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr23_+_26142807 | 0.11 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr10_+_35352435 | 0.11 |
ENSDART00000123936
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr5_+_51443009 | 0.11 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr6_+_612594 | 0.11 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr12_-_36521767 | 0.11 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr8_+_4798158 | 0.11 |
ENSDART00000031650
|
hsp70l
|
heat shock cognate 70-kd protein, like |
| chr22_-_621888 | 0.10 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr14_+_20893065 | 0.10 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr25_+_30131055 | 0.10 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr10_+_38708099 | 0.10 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr22_-_26852516 | 0.10 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr3_-_36209936 | 0.10 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr23_+_4022620 | 0.10 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr8_-_4100365 | 0.10 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr3_-_8012847 | 0.10 |
ENSDART00000169673
|
si:ch211-175l6.1
|
si:ch211-175l6.1 |
| chr2_-_17114852 | 0.10 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr24_-_33308045 | 0.10 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr10_-_36808348 | 0.10 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr7_-_54679595 | 0.09 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr1_+_41588170 | 0.09 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr6_-_436658 | 0.09 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr10_-_17587832 | 0.09 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr23_-_31372639 | 0.09 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr3_-_23513155 | 0.09 |
ENSDART00000170200
|
BX682558.1
|
|
| chr21_+_26522571 | 0.09 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr12_+_13205955 | 0.09 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr3_-_17716322 | 0.09 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr17_-_36529016 | 0.09 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr7_+_4523051 | 0.09 |
ENSDART00000140860
ENSDART00000143778 |
si:dkey-83f18.10
|
si:dkey-83f18.10 |
| chr5_-_42083363 | 0.09 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr11_-_37997419 | 0.09 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr7_-_71585065 | 0.09 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr23_+_20431140 | 0.09 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr9_-_14137295 | 0.08 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr11_-_31226578 | 0.08 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr5_+_26795773 | 0.08 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr14_+_23709134 | 0.08 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_23596391 | 0.08 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr17_+_42274825 | 0.08 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr3_+_8224622 | 0.08 |
ENSDART00000138670
|
si:ch73-379f5.5
|
si:ch73-379f5.5 |
| chr4_+_3980247 | 0.08 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr23_+_36063599 | 0.08 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr8_-_30338872 | 0.08 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr2_-_36925561 | 0.08 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr16_-_46619967 | 0.08 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr22_-_31020690 | 0.08 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr5_+_22393501 | 0.08 |
ENSDART00000185276
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr2_+_36112273 | 0.08 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr8_+_48942470 | 0.08 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr8_-_25033681 | 0.08 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr3_+_12718100 | 0.08 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr9_+_14152211 | 0.08 |
ENSDART00000148055
|
si:ch211-67e16.11
|
si:ch211-67e16.11 |
| chr25_-_27541621 | 0.07 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr4_+_9478500 | 0.07 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr4_+_76705830 | 0.07 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr17_+_6563307 | 0.07 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr6_+_22326624 | 0.07 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr9_+_30478768 | 0.07 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr3_+_22377312 | 0.07 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr10_-_10018120 | 0.07 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr22_-_16755885 | 0.07 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr4_+_15968483 | 0.06 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr16_-_30903930 | 0.06 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr21_+_28958471 | 0.06 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_+_29209926 | 0.06 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr22_+_6149121 | 0.06 |
ENSDART00000134067
|
si:dkey-19a16.5
|
si:dkey-19a16.5 |
| chr16_+_14029283 | 0.06 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr22_-_17653143 | 0.06 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr22_-_3299100 | 0.06 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr12_-_10505986 | 0.06 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr18_+_15132112 | 0.06 |
ENSDART00000099764
|
zgc:153031
|
zgc:153031 |
| chr14_-_36437249 | 0.06 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr22_-_19504690 | 0.06 |
ENSDART00000166296
|
si:dkey-78l4.11
|
si:dkey-78l4.11 |
| chr20_-_38446891 | 0.06 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr13_+_48359573 | 0.06 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr10_-_43718914 | 0.05 |
ENSDART00000189277
|
cetn3
|
centrin 3 |
| chr22_-_24984733 | 0.05 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr19_-_7272921 | 0.05 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr8_+_20140321 | 0.05 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr17_-_46457622 | 0.05 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr21_-_43606502 | 0.05 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr21_-_4922981 | 0.05 |
ENSDART00000191819
ENSDART00000113556 ENSDART00000193269 ENSDART00000182961 |
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr4_-_15103646 | 0.05 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr21_-_30408775 | 0.05 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr7_+_3872189 | 0.05 |
ENSDART00000064221
|
si:dkey-88n24.5
|
si:dkey-88n24.5 |
| chr2_+_11685742 | 0.05 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr9_+_44430974 | 0.05 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr21_-_4923427 | 0.05 |
ENSDART00000181489
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr11_-_18107447 | 0.05 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr2_+_27010439 | 0.05 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr11_-_35975026 | 0.05 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr4_+_71086017 | 0.05 |
ENSDART00000159113
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr19_-_48019366 | 0.05 |
ENSDART00000169429
|
si:ch1073-205c8.5
|
si:ch1073-205c8.5 |
| chr14_-_31693148 | 0.05 |
ENSDART00000172958
|
map7d3
|
MAP7 domain containing 3 |
| chr6_-_18618106 | 0.05 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr1_-_12397258 | 0.05 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr13_-_42536642 | 0.05 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr7_+_26998169 | 0.05 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr23_-_38497705 | 0.05 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr4_+_15899668 | 0.05 |
ENSDART00000101592
|
col7a1l
|
collagen type VII alpha 1-like |
| chr8_+_18830759 | 0.05 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr21_-_35325466 | 0.05 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr11_+_21587950 | 0.05 |
ENSDART00000162251
|
FAM72B
|
zgc:101564 |
| chr22_-_14115292 | 0.04 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr9_+_54695981 | 0.04 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr17_+_44756247 | 0.04 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr4_-_12930086 | 0.04 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr17_+_28340138 | 0.04 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr5_+_9360394 | 0.04 |
ENSDART00000124642
|
FP236810.2
|
|
| chr7_+_15871156 | 0.04 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr11_+_45110865 | 0.04 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr13_+_15701849 | 0.04 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr7_+_8543317 | 0.04 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr12_+_36416173 | 0.04 |
ENSDART00000190278
|
unk
|
unkempt family zinc finger |
| chr21_-_39327223 | 0.04 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr16_+_24626448 | 0.04 |
ENSDART00000153591
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr4_-_890220 | 0.04 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr20_+_25712276 | 0.04 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr1_+_22174251 | 0.04 |
ENSDART00000137429
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr12_+_7491690 | 0.04 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr17_-_51224800 | 0.04 |
ENSDART00000150089
|
psen1
|
presenilin 1 |
| chr22_-_29689485 | 0.04 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr4_+_26056548 | 0.04 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr7_+_24496894 | 0.04 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr6_-_35738836 | 0.04 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr4_+_57099307 | 0.04 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr20_+_25711718 | 0.04 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr17_-_19535328 | 0.04 |
ENSDART00000077809
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr9_+_3283608 | 0.04 |
ENSDART00000192275
|
hat1
|
histone acetyltransferase 1 |
| chr3_-_18737126 | 0.04 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr6_-_19023468 | 0.04 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr10_+_7593185 | 0.04 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr4_+_20566371 | 0.04 |
ENSDART00000127576
|
BX248410.1
|
|
| chr16_+_9762261 | 0.04 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.3 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0072378 | platelet aggregation(GO:0070527) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.1 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |