Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
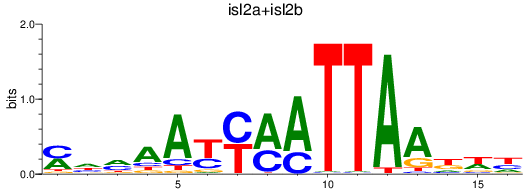
Results for isl2a+isl2b
Z-value: 0.83
Transcription factors associated with isl2a+isl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl2a
|
ENSDARG00000003971 | ISL LIM homeobox 2a |
|
isl2b
|
ENSDARG00000053499 | ISL LIM homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl2b | dr11_v1_chr7_+_29992889_29992889 | 0.52 | 3.7e-01 | Click! |
| isl2a | dr11_v1_chr25_-_32751982_32751982 | -0.38 | 5.3e-01 | Click! |
Activity profile of isl2a+isl2b motif
Sorted Z-values of isl2a+isl2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_24573721 | 0.66 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr13_-_2010191 | 0.59 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr2_-_38284648 | 0.45 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr15_-_2640966 | 0.42 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr21_+_11385031 | 0.39 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr8_-_50147948 | 0.36 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr5_+_58550291 | 0.36 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr22_-_16154771 | 0.30 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr22_-_17482282 | 0.30 |
ENSDART00000105453
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr25_+_22320738 | 0.29 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr6_+_39905021 | 0.28 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr3_+_28939759 | 0.27 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr15_+_46356879 | 0.27 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr19_-_5769728 | 0.27 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr5_+_69785990 | 0.26 |
ENSDART00000162057
ENSDART00000166893 |
kmt5ab
|
lysine methyltransferase 5Ab |
| chr21_-_10886709 | 0.26 |
ENSDART00000134408
|
si:dkey-277m11.2
|
si:dkey-277m11.2 |
| chr7_+_60079302 | 0.25 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr21_+_25236297 | 0.24 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr15_+_46357080 | 0.24 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr21_-_44081540 | 0.24 |
ENSDART00000130833
|
FO704810.1
|
|
| chr5_+_69786215 | 0.23 |
ENSDART00000165956
|
kmt5ab
|
lysine methyltransferase 5Ab |
| chr12_-_4532066 | 0.22 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr1_-_25966068 | 0.22 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr3_+_3681116 | 0.22 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr15_-_46779934 | 0.22 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr23_-_31645760 | 0.21 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_+_44956660 | 0.21 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr1_+_52792439 | 0.21 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr20_-_33507458 | 0.21 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr8_+_24747865 | 0.21 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr11_-_1400507 | 0.20 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr16_-_48400639 | 0.20 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr10_+_29850330 | 0.19 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr7_-_66868543 | 0.19 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr23_-_16485190 | 0.19 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr16_-_45094599 | 0.19 |
ENSDART00000155479
|
si:rp71-77l1.1
|
si:rp71-77l1.1 |
| chr19_-_5125663 | 0.18 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr22_-_10165446 | 0.18 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr15_-_21702317 | 0.18 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr13_-_42560662 | 0.18 |
ENSDART00000124898
|
CR792417.1
|
|
| chr22_-_22147375 | 0.18 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr11_-_10456553 | 0.17 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr11_-_10456387 | 0.17 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr2_-_10877765 | 0.17 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr16_+_23303859 | 0.17 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr21_+_11749097 | 0.17 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr18_-_48547564 | 0.16 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr16_-_16761164 | 0.16 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr3_+_26342768 | 0.16 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr22_-_10470663 | 0.16 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr6_-_52788213 | 0.16 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr11_+_31324335 | 0.16 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_-_20403845 | 0.16 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr14_-_4145594 | 0.16 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr20_+_41021054 | 0.16 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr2_+_56657804 | 0.16 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr1_+_55608520 | 0.16 |
ENSDART00000152307
|
adgre18
|
adhesion G protein-coupled receptor E18 |
| chr18_+_30847237 | 0.16 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr18_+_13275735 | 0.15 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr22_-_17482810 | 0.15 |
ENSDART00000171293
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr13_+_646700 | 0.15 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr11_+_24002503 | 0.15 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr12_+_17154655 | 0.15 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr25_-_21031007 | 0.15 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr3_-_25268751 | 0.15 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr18_-_15932704 | 0.15 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr9_-_394088 | 0.15 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr20_-_5291012 | 0.14 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr23_+_32426954 | 0.14 |
ENSDART00000192529
ENSDART00000172313 |
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr6_-_35046735 | 0.14 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr6_+_54538948 | 0.14 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr23_+_32406461 | 0.14 |
ENSDART00000179878
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr17_-_14815557 | 0.14 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr6_-_54486410 | 0.13 |
ENSDART00000064679
|
CR847944.1
|
|
| chr23_-_33350990 | 0.13 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr1_-_9641845 | 0.13 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr24_+_16985181 | 0.13 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr2_+_20605925 | 0.13 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr22_+_19552987 | 0.13 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr24_-_26310854 | 0.13 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr2_+_36112273 | 0.13 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr20_+_11731039 | 0.12 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr11_+_24001993 | 0.12 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr1_+_51721851 | 0.12 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr3_+_32553714 | 0.12 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr12_+_16168342 | 0.12 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr23_-_3511630 | 0.12 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr6_-_12588044 | 0.12 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr5_-_42894068 | 0.12 |
ENSDART00000169606
|
cxcl11.1
|
chemokine (C-X-C motif) ligand 11, duplicate 1 |
| chr10_-_34916208 | 0.12 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr9_+_24008879 | 0.12 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr12_-_28363111 | 0.12 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr21_-_35419486 | 0.11 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr15_+_34988148 | 0.11 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr3_+_14388010 | 0.11 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr3_-_54846444 | 0.11 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr21_-_19314618 | 0.11 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr7_+_59020972 | 0.11 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr3_-_19368435 | 0.11 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr22_-_9183944 | 0.11 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr24_+_13869092 | 0.10 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_+_11660839 | 0.10 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr23_+_36460239 | 0.10 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr2_-_59205393 | 0.10 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr2_+_19633493 | 0.10 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr19_+_16015881 | 0.10 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr3_+_23029934 | 0.10 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr19_+_16016038 | 0.10 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr23_+_39695827 | 0.10 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr8_-_53044300 | 0.10 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_-_47323267 | 0.09 |
ENSDART00000190077
|
CU138512.1
|
|
| chr9_+_48761455 | 0.09 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr19_+_31904836 | 0.09 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr23_-_16484383 | 0.09 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr13_+_9468535 | 0.09 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr19_+_1688727 | 0.09 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_4245902 | 0.09 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr13_-_9450210 | 0.08 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr24_+_19415124 | 0.08 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr22_-_36530902 | 0.08 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr24_-_3477103 | 0.08 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr11_-_29737088 | 0.08 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr5_-_42083363 | 0.08 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr15_+_25438714 | 0.08 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr12_-_6880694 | 0.08 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr19_+_9186175 | 0.08 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr3_+_23029484 | 0.08 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr2_+_19522082 | 0.08 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr6_-_49510553 | 0.08 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr10_-_17083180 | 0.08 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr13_+_9046473 | 0.08 |
ENSDART00000136557
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr2_-_48512450 | 0.08 |
ENSDART00000160132
|
CR391991.1
|
|
| chr16_-_17072440 | 0.08 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr22_-_12745589 | 0.07 |
ENSDART00000136841
|
plcd4a
|
phospholipase C, delta 4a |
| chr8_-_7475917 | 0.07 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr3_-_13068189 | 0.07 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_-_12407106 | 0.07 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr20_+_25586099 | 0.07 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr7_+_66822229 | 0.07 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr8_-_13046089 | 0.07 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr13_-_9467944 | 0.07 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr14_+_21722235 | 0.07 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr17_-_2834764 | 0.07 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr5_+_55225497 | 0.06 |
ENSDART00000144087
|
tmc2a
|
transmembrane channel-like 2a |
| chr6_+_9175886 | 0.06 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr13_-_33639050 | 0.06 |
ENSDART00000133073
|
rrbp1a
|
ribosome binding protein 1a |
| chr2_-_19520690 | 0.06 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr25_+_13406069 | 0.06 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr23_-_18024543 | 0.06 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr4_-_77218637 | 0.06 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr8_+_31717175 | 0.06 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr10_+_31248036 | 0.06 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr1_+_45002971 | 0.06 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr11_-_45138857 | 0.06 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr13_-_30996072 | 0.06 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr2_-_22927958 | 0.06 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr7_-_7692723 | 0.05 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr21_+_45841731 | 0.05 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr16_-_10223741 | 0.05 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr22_-_34995333 | 0.05 |
ENSDART00000110900
|
kcnip2
|
Kv channel interacting protein 2 |
| chr3_-_25119839 | 0.05 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr19_+_7424347 | 0.05 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr7_-_7692992 | 0.05 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr8_+_31716872 | 0.05 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr13_-_9045879 | 0.05 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr3_-_59297532 | 0.05 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr24_+_12894282 | 0.05 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr9_+_32069989 | 0.05 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr7_+_20512419 | 0.05 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr2_+_10771787 | 0.05 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr24_-_12770357 | 0.05 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr9_+_51147380 | 0.04 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr17_-_24353462 | 0.04 |
ENSDART00000191021
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr6_+_12527725 | 0.04 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr22_-_17844117 | 0.04 |
ENSDART00000159363
|
BX908731.1
|
|
| chr7_-_48665305 | 0.04 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr10_-_40964739 | 0.04 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr5_-_19861766 | 0.04 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr12_+_18899396 | 0.04 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr2_+_19578446 | 0.04 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr8_+_52637507 | 0.04 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr21_+_43172506 | 0.03 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr19_+_24872159 | 0.03 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr6_+_7533601 | 0.03 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr19_+_45962016 | 0.03 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr2_+_10878406 | 0.03 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr13_+_7049823 | 0.03 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr2_-_19520324 | 0.03 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr3_+_38540411 | 0.03 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr18_+_3579829 | 0.03 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr24_+_2495197 | 0.03 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr4_-_49582758 | 0.03 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr6_+_9204099 | 0.03 |
ENSDART00000150167
ENSDART00000115394 |
tbx19
|
T-box 19 |
| chr8_-_22273819 | 0.03 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr18_-_8398972 | 0.03 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr2_-_7845110 | 0.02 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr24_-_6078222 | 0.02 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr17_+_12285285 | 0.02 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr17_+_43595692 | 0.02 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr1_-_52210950 | 0.02 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl2a+isl2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.4 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.0 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |