Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for jun
Z-value: 1.03
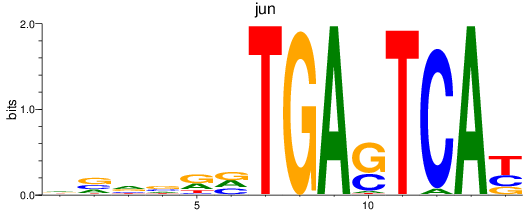
Transcription factors associated with jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr11_v1_chr20_+_15552657_15552657 | -0.83 | 7.9e-02 | Click! |
Activity profile of jun motif
Sorted Z-values of jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23403602 | 0.85 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr20_+_28364742 | 0.76 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr13_-_33134611 | 0.74 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr7_+_69019851 | 0.74 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr21_+_25765734 | 0.67 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr5_+_45677781 | 0.61 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr15_+_963292 | 0.58 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr8_+_5024468 | 0.56 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr25_+_18583877 | 0.53 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_+_14005111 | 0.51 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr8_-_38201415 | 0.50 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr19_-_7420867 | 0.49 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr23_+_17522867 | 0.48 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr20_+_26880668 | 0.47 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr19_+_48359259 | 0.47 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr13_+_23988442 | 0.46 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr10_+_28428222 | 0.45 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr25_+_10410620 | 0.43 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr16_-_36834505 | 0.43 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr25_-_22191733 | 0.42 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr2_+_25839940 | 0.42 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr21_+_26612777 | 0.41 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr1_-_25679339 | 0.41 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr1_-_25177086 | 0.40 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr1_-_59077650 | 0.39 |
ENSDART00000043516
|
MFAP4 (1 of many)
|
si:zfos-2330d3.1 |
| chr23_-_1017605 | 0.39 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr10_+_32050906 | 0.39 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr8_+_8671229 | 0.39 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr6_+_56141852 | 0.38 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_+_24089334 | 0.38 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr9_+_33216945 | 0.38 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr7_+_25033924 | 0.37 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr25_-_13408760 | 0.37 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr25_+_29474583 | 0.37 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr10_+_26747755 | 0.36 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr1_-_58913813 | 0.36 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr5_-_57480660 | 0.36 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr3_+_32118670 | 0.36 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr10_-_3295197 | 0.35 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr9_+_33217243 | 0.35 |
ENSDART00000053061
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr22_-_24858042 | 0.34 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr13_+_829585 | 0.34 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr23_-_1017428 | 0.33 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr23_+_25856541 | 0.33 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr6_+_51713076 | 0.33 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr18_+_47313899 | 0.33 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr17_-_53359028 | 0.32 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr16_-_21785261 | 0.32 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr10_-_22095505 | 0.31 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr4_+_77948970 | 0.30 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_+_9037650 | 0.30 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr22_+_19218733 | 0.30 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr15_-_46779934 | 0.30 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr19_-_5332784 | 0.29 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr8_-_18667693 | 0.29 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr18_+_47313715 | 0.28 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr3_-_26017592 | 0.28 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr22_-_24297510 | 0.28 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr24_-_23839647 | 0.28 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr3_-_48259289 | 0.28 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr6_-_43449013 | 0.28 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr24_+_30215475 | 0.28 |
ENSDART00000164717
|
si:ch73-358j7.2
|
si:ch73-358j7.2 |
| chr8_+_6576940 | 0.28 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr23_-_45398622 | 0.27 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr19_-_27570333 | 0.27 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr14_+_30774032 | 0.27 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr16_+_46410520 | 0.27 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr20_+_35445462 | 0.27 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr6_-_49063085 | 0.27 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr3_-_26017831 | 0.27 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr19_-_27564458 | 0.27 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr1_+_54655160 | 0.27 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr4_+_16715267 | 0.26 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr7_+_33172066 | 0.26 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr23_+_4324625 | 0.26 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr20_-_26531850 | 0.26 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr20_+_53522059 | 0.26 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr3_+_31680592 | 0.26 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr22_+_19538626 | 0.26 |
ENSDART00000189174
|
BX936298.2
|
|
| chr25_-_7753207 | 0.25 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr2_+_38731696 | 0.25 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr3_+_21189766 | 0.25 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr3_+_3598555 | 0.25 |
ENSDART00000191152
|
CR589947.3
|
|
| chr24_-_33366188 | 0.24 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr2_-_898899 | 0.24 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr22_-_5839686 | 0.24 |
ENSDART00000145406
|
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr14_+_30774515 | 0.24 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr21_+_33459524 | 0.24 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr11_-_23501467 | 0.24 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_-_13518381 | 0.23 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr24_+_24726956 | 0.23 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr6_-_43028896 | 0.23 |
ENSDART00000149977
|
glyctk
|
glycerate kinase |
| chr8_+_39511932 | 0.22 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr25_-_12824939 | 0.22 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr20_-_26532167 | 0.22 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr3_-_3448095 | 0.22 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr8_-_23573084 | 0.22 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr25_+_29474982 | 0.22 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr14_+_1240235 | 0.22 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr1_+_59073436 | 0.22 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr17_-_15600455 | 0.22 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr18_+_6558338 | 0.22 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr2_+_25839650 | 0.22 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr5_-_20123002 | 0.21 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr1_+_57041549 | 0.21 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr4_+_9508505 | 0.21 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr2_+_25840463 | 0.21 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr15_-_38129845 | 0.21 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr16_+_41517188 | 0.21 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr20_+_7084154 | 0.21 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr13_-_37649595 | 0.21 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr25_+_23336310 | 0.21 |
ENSDART00000156457
|
ptprjb.2
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 2 |
| chr15_+_3766101 | 0.21 |
ENSDART00000042580
ENSDART00000112698 ENSDART00000187035 ENSDART00000165571 ENSDART00000121752 |
RNF14 (1 of many)
|
zmp:0000000524 |
| chr7_-_8470860 | 0.20 |
ENSDART00000172793
|
loc564481
|
hypothetical protein LOC564481 |
| chr1_+_59090743 | 0.20 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr1_-_59252973 | 0.20 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr1_+_59090972 | 0.20 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr9_+_33417969 | 0.20 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr4_+_25181572 | 0.20 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr3_+_46762703 | 0.20 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_71389375 | 0.20 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr2_+_36109002 | 0.20 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr5_+_42141917 | 0.20 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr16_+_49005321 | 0.20 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr20_-_35578435 | 0.20 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr18_+_15758375 | 0.20 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr16_-_40373836 | 0.19 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr3_+_22375596 | 0.19 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr9_+_8380728 | 0.19 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr10_-_44560165 | 0.19 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr22_-_817479 | 0.19 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr14_-_22100118 | 0.19 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr8_-_11170114 | 0.19 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr13_-_15982707 | 0.18 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_+_77971104 | 0.18 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr19_+_24575077 | 0.18 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr10_-_44482911 | 0.18 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr6_+_48348415 | 0.18 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr6_-_12588044 | 0.18 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr23_-_36316352 | 0.18 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr19_+_15441022 | 0.17 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr24_-_23998897 | 0.17 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr25_-_29072162 | 0.17 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr13_-_37122217 | 0.17 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr2_-_37277626 | 0.17 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr17_+_45413324 | 0.17 |
ENSDART00000124911
|
ezra
|
ezrin a |
| chr8_+_32406885 | 0.17 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr5_-_1047504 | 0.17 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr20_-_25626198 | 0.17 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr11_+_37905630 | 0.16 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr19_+_7567763 | 0.16 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr22_+_19247255 | 0.16 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr2_-_985417 | 0.16 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr21_+_20901505 | 0.16 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr22_+_1092479 | 0.16 |
ENSDART00000170119
|
guca1e
|
guanylate cyclase activator 1e |
| chr24_-_30275204 | 0.16 |
ENSDART00000164187
|
snx7
|
sorting nexin 7 |
| chr6_+_42475730 | 0.16 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr3_-_15734358 | 0.16 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr1_+_14253118 | 0.16 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr18_+_8320165 | 0.16 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr2_+_30379650 | 0.16 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr8_+_28695914 | 0.16 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr7_+_34786591 | 0.16 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr7_+_34549377 | 0.15 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr7_-_3630019 | 0.15 |
ENSDART00000138164
ENSDART00000064277 |
si:dkey-192d15.3
|
si:dkey-192d15.3 |
| chr22_+_19405517 | 0.15 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr22_+_5532003 | 0.15 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr25_-_12824656 | 0.15 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr5_+_50953240 | 0.15 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr2_+_47945359 | 0.15 |
ENSDART00000098054
|
ftr19
|
finTRIM family, member 19 |
| chr10_+_38775408 | 0.15 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr10_+_38775959 | 0.15 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr3_+_1179601 | 0.15 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr4_-_57572054 | 0.15 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr22_+_4442473 | 0.15 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr3_-_15734530 | 0.15 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr20_+_9683994 | 0.15 |
ENSDART00000053831
|
ptger2b
|
prostaglandin E receptor 2b (subtype EP2) |
| chr19_-_42573219 | 0.15 |
ENSDART00000126021
ENSDART00000133695 ENSDART00000131558 |
zgc:103438
|
zgc:103438 |
| chr13_+_10621257 | 0.15 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr8_-_44463985 | 0.15 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr23_+_25232711 | 0.14 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr12_+_20587179 | 0.14 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr4_+_77933084 | 0.14 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_+_24251141 | 0.14 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr20_+_25625872 | 0.14 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr22_+_9862243 | 0.14 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr15_+_714203 | 0.14 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr23_-_4705110 | 0.14 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr8_-_20243389 | 0.14 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr22_+_19365220 | 0.14 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr21_+_20903244 | 0.14 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr20_-_38746889 | 0.14 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr19_-_27564980 | 0.14 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr4_+_47656992 | 0.14 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr5_-_20195350 | 0.14 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr6_-_24143923 | 0.14 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr5_+_22393501 | 0.14 |
ENSDART00000185276
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr5_-_30615901 | 0.14 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr19_+_15443063 | 0.14 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_+_19687217 | 0.14 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.4 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.8 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.6 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 0.2 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.2 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.2 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.6 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.5 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.4 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.5 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.5 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.4 | GO:0072378 | platelet aggregation(GO:0070527) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:1905208 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0007620 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0032355 | response to estradiol(GO:0032355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.2 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.2 | GO:0010853 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |