Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for jund_batf
Z-value: 1.06
Transcription factors associated with jund_batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
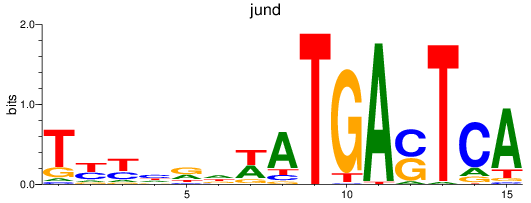
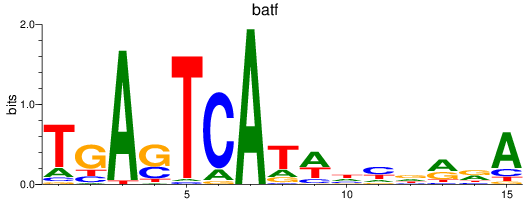
jund
|
ENSDARG00000067850 | JunD proto-oncogene, AP-1 transcription factor subunit |
|
batf
|
ENSDARG00000011818 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jund | dr11_v1_chr2_-_56131312_56131312 | -0.92 | 2.7e-02 | Click! |
| batf | dr11_v1_chr20_+_46572550_46572550 | -0.50 | 3.9e-01 | Click! |
Activity profile of jund_batf motif
Sorted Z-values of jund_batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_56176976 | 1.46 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr7_+_25036188 | 1.39 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr3_-_26017592 | 1.25 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr13_-_20381485 | 1.11 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr3_-_26017831 | 1.09 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr10_+_28428222 | 0.95 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr16_-_13818061 | 0.93 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr8_-_38201415 | 0.84 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_+_38173960 | 0.81 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr1_+_135903 | 0.72 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr22_+_19218733 | 0.70 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr4_-_13518381 | 0.69 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr9_+_38645136 | 0.67 |
ENSDART00000135505
|
slc12a8
|
solute carrier family 12, member 8 |
| chr25_+_10410620 | 0.64 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr9_+_38644976 | 0.63 |
ENSDART00000133849
ENSDART00000135774 |
slc12a8
|
solute carrier family 12, member 8 |
| chr5_+_66170479 | 0.61 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr14_+_30774032 | 0.60 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr14_+_30774515 | 0.59 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr21_+_25765734 | 0.59 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr19_+_37118547 | 0.58 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr6_-_43449013 | 0.57 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr15_-_21877726 | 0.57 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr20_+_34868933 | 0.55 |
ENSDART00000153006
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr25_+_18583877 | 0.54 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_+_55149001 | 0.52 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr7_+_29509255 | 0.52 |
ENSDART00000076172
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr11_-_2478374 | 0.52 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr23_-_4975452 | 0.51 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr22_+_19247255 | 0.50 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr1_+_14253118 | 0.49 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr22_+_16497670 | 0.46 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr8_-_36554675 | 0.45 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr14_+_30774894 | 0.42 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr22_+_19405517 | 0.42 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr18_-_30499489 | 0.41 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr23_+_30967686 | 0.41 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr6_+_598669 | 0.39 |
ENSDART00000151009
|
si:ch73-379f7.4
|
si:ch73-379f7.4 |
| chr25_+_29474982 | 0.39 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr10_+_40700311 | 0.38 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr23_-_1017428 | 0.37 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr1_-_58913813 | 0.34 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr1_-_35694978 | 0.32 |
ENSDART00000136157
|
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr12_+_46634736 | 0.32 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr11_-_40101246 | 0.31 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr20_-_33704753 | 0.31 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr4_-_72476526 | 0.30 |
ENSDART00000174153
|
CR788316.1
|
|
| chr13_-_9335891 | 0.30 |
ENSDART00000080637
|
BX901922.1
|
|
| chr19_+_10527228 | 0.30 |
ENSDART00000091918
|
si:ch73-160i9.3
|
si:ch73-160i9.3 |
| chr24_-_38192003 | 0.30 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr15_+_15771418 | 0.29 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr4_+_42175261 | 0.28 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr16_-_25608453 | 0.28 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr12_+_16132612 | 0.28 |
ENSDART00000152550
|
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr22_+_28337204 | 0.28 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr5_-_41531629 | 0.27 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr3_-_39488482 | 0.26 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr16_-_24832038 | 0.26 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr9_-_1978090 | 0.26 |
ENSDART00000082344
|
hoxd11a
|
homeobox D11a |
| chr23_-_1017605 | 0.26 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr19_-_5669122 | 0.26 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr1_+_57741577 | 0.26 |
ENSDART00000192673
|
LO018597.1
|
|
| chr5_+_26799165 | 0.25 |
ENSDART00000145736
|
tcn2
|
transcobalamin II |
| chr11_+_45219558 | 0.25 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr4_+_1530287 | 0.25 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr7_+_34786591 | 0.25 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr3_-_39488639 | 0.25 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr25_-_29072162 | 0.25 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_+_1911814 | 0.25 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr12_+_41348969 | 0.25 |
ENSDART00000171352
|
si:ch211-27e6.1
|
si:ch211-27e6.1 |
| chr12_-_21684197 | 0.25 |
ENSDART00000152999
ENSDART00000153109 ENSDART00000148698 |
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr22_-_817479 | 0.25 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr3_-_16493528 | 0.24 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr17_-_6536466 | 0.24 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr22_+_36914636 | 0.23 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr22_+_19266995 | 0.23 |
ENSDART00000133995
ENSDART00000144963 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr14_-_976912 | 0.23 |
ENSDART00000114053
|
arsia
|
arylsulfatase family, member Ia |
| chr9_+_14010823 | 0.23 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr13_-_5257303 | 0.23 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr25_+_29474583 | 0.22 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr19_+_42142381 | 0.22 |
ENSDART00000129915
|
kcnq4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr12_-_25294096 | 0.22 |
ENSDART00000183398
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr16_+_8695595 | 0.22 |
ENSDART00000173249
|
si:cabz01093077.1
|
si:cabz01093077.1 |
| chr2_+_27855102 | 0.22 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr1_-_43897831 | 0.21 |
ENSDART00000048225
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr7_+_6317866 | 0.20 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr20_+_572037 | 0.20 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr25_-_19666107 | 0.20 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr19_+_40856807 | 0.20 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr23_+_19655301 | 0.20 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr15_+_17722054 | 0.20 |
ENSDART00000191390
ENSDART00000169550 |
si:ch211-213d14.1
|
si:ch211-213d14.1 |
| chr7_+_44802353 | 0.20 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr14_+_10461264 | 0.20 |
ENSDART00000081095
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr6_+_56141852 | 0.20 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_+_68088529 | 0.20 |
ENSDART00000159155
|
znf1096
|
zinc finger protein 1096 |
| chr22_+_9294336 | 0.20 |
ENSDART00000193694
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr14_+_3507326 | 0.19 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr3_-_4760384 | 0.19 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr22_+_5851122 | 0.19 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr1_+_59088205 | 0.19 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr2_-_24068848 | 0.19 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr16_+_19029297 | 0.19 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr9_+_33334501 | 0.19 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr20_-_2134620 | 0.18 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr3_-_34070451 | 0.18 |
ENSDART00000151458
|
ighv8-3
|
immunoglobulin heavy variable 8-3 |
| chr5_+_24089334 | 0.18 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr1_-_11606903 | 0.18 |
ENSDART00000136093
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr17_-_6536305 | 0.17 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr7_-_60351876 | 0.17 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr21_-_21530868 | 0.17 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr9_-_24244383 | 0.17 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr5_-_57480660 | 0.17 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr22_+_19365220 | 0.16 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr6_-_43028896 | 0.16 |
ENSDART00000149977
|
glyctk
|
glycerate kinase |
| chr4_-_60780423 | 0.16 |
ENSDART00000162632
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr8_+_25034544 | 0.16 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr14_-_22100118 | 0.16 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr3_-_21402279 | 0.16 |
ENSDART00000164513
|
CT573446.1
|
|
| chr7_-_13906409 | 0.16 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr3_+_30968176 | 0.16 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr2_-_24069331 | 0.16 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr2_-_6519017 | 0.15 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr5_-_25645593 | 0.15 |
ENSDART00000044471
|
tmc2b
|
transmembrane channel-like 2b |
| chr15_-_29586747 | 0.15 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr3_-_16289826 | 0.15 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_+_42141917 | 0.15 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr8_-_7603700 | 0.15 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr24_+_7800486 | 0.15 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr3_-_61185746 | 0.15 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr7_-_35409027 | 0.14 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr21_+_33459524 | 0.14 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr4_+_54899568 | 0.14 |
ENSDART00000162786
|
si:dkey-56m15.8
|
si:dkey-56m15.8 |
| chr25_-_35169303 | 0.14 |
ENSDART00000193240
|
ano9a
|
anoctamin 9a |
| chr7_-_35408618 | 0.14 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr4_+_34121902 | 0.14 |
ENSDART00000170225
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr16_+_49005321 | 0.14 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr8_+_13389115 | 0.14 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr19_-_27395531 | 0.14 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr4_-_50930346 | 0.14 |
ENSDART00000184245
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr5_-_24270989 | 0.13 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr14_-_34074510 | 0.13 |
ENSDART00000172753
|
itk
|
IL2 inducible T cell kinase |
| chr4_-_36476889 | 0.13 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr5_-_30984010 | 0.13 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr7_-_6357952 | 0.13 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr25_-_11026907 | 0.13 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr21_+_10790680 | 0.12 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr8_-_44463985 | 0.12 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr8_-_3413139 | 0.12 |
ENSDART00000182673
ENSDART00000166741 ENSDART00000169430 ENSDART00000170478 |
FUT9 (1 of many)
FUT9 (1 of many)
fut9b
|
zgc:103510 zgc:165519 fucosyltransferase 9b |
| chr2_-_49997055 | 0.12 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr3_+_16976095 | 0.12 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr10_+_7718156 | 0.12 |
ENSDART00000189101
|
ggcx
|
gamma-glutamyl carboxylase |
| chr2_+_36608387 | 0.12 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr22_-_26595027 | 0.12 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr1_+_58442694 | 0.12 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr4_+_47436126 | 0.12 |
ENSDART00000157555
|
si:dkey-124l13.1
|
si:dkey-124l13.1 |
| chr11_+_21050326 | 0.11 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr20_+_53474963 | 0.11 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr22_-_12746539 | 0.11 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr8_-_25034411 | 0.11 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr20_-_34127415 | 0.11 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr16_+_16824678 | 0.11 |
ENSDART00000172862
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr9_+_2499627 | 0.11 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr4_+_64549970 | 0.10 |
ENSDART00000167846
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr16_+_18974064 | 0.10 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr4_-_17793152 | 0.10 |
ENSDART00000134080
|
mybpc1
|
myosin binding protein C, slow type |
| chr25_-_15504559 | 0.10 |
ENSDART00000139294
|
BX323543.5
|
|
| chr10_+_31248036 | 0.10 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr25_-_15496485 | 0.10 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr8_+_36554816 | 0.10 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr1_-_47122058 | 0.10 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr7_-_17591007 | 0.10 |
ENSDART00000171023
|
CU672228.1
|
|
| chr4_-_75899294 | 0.10 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr5_+_50953240 | 0.09 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr4_-_42397126 | 0.09 |
ENSDART00000162437
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr24_+_26658132 | 0.09 |
ENSDART00000183081
|
FQ378040.1
|
|
| chr4_+_51564997 | 0.09 |
ENSDART00000186119
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr6_+_59832786 | 0.09 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr5_-_43959972 | 0.09 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr24_+_31361407 | 0.09 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr4_+_54645654 | 0.09 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr2_+_30379650 | 0.09 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr16_-_46567136 | 0.09 |
ENSDART00000159180
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr21_-_2415808 | 0.08 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr19_+_26072624 | 0.08 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr16_+_46410520 | 0.08 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr4_-_71913556 | 0.08 |
ENSDART00000167608
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr5_+_55265274 | 0.08 |
ENSDART00000131060
|
pcsk5a
|
proprotein convertase subtilisin/kexin type 5a |
| chr4_-_33071267 | 0.08 |
ENSDART00000186314
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr14_-_32884138 | 0.08 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr25_+_22587306 | 0.08 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr4_-_50434519 | 0.08 |
ENSDART00000150372
|
znf1061
|
zinc finger protein 1061 |
| chr4_+_57194439 | 0.08 |
ENSDART00000075265
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr25_-_15512819 | 0.08 |
ENSDART00000142684
|
si:dkeyp-67e1.2
|
si:dkeyp-67e1.2 |
| chr23_-_18024543 | 0.08 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr4_-_39265279 | 0.08 |
ENSDART00000164912
|
si:ch73-236c18.2
|
si:ch73-236c18.2 |
| chr18_-_15771551 | 0.08 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr20_-_32658305 | 0.07 |
ENSDART00000140622
ENSDART00000152909 |
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr10_+_31344227 | 0.07 |
ENSDART00000184470
|
AL845320.2
|
|
| chr13_-_42749916 | 0.07 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr22_-_20289948 | 0.07 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr12_+_46740584 | 0.07 |
ENSDART00000171563
|
plaub
|
plasminogen activator, urokinase b |
Network of associatons between targets according to the STRING database.
First level regulatory network of jund_batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 0.5 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 0.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 1.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.2 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.5 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.6 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 1.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.8 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |