Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
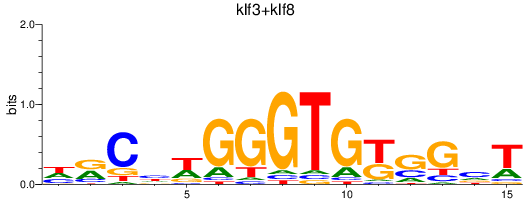
Results for klf3+klf8
Z-value: 0.79
Transcription factors associated with klf3+klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf3
|
ENSDARG00000015495 | Kruppel-like factor 3 (basic) |
|
klf8
|
ENSDARG00000060661 | Kruppel-like factor 8 |
|
klf3
|
ENSDARG00000114143 | Kruppel-like factor 3 (basic) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf8 | dr11_v1_chr21_+_38089036_38089036 | -0.73 | 1.6e-01 | Click! |
| klf3 | dr11_v1_chr1_-_18615063_18615063 | 0.06 | 9.2e-01 | Click! |
Activity profile of klf3+klf8 motif
Sorted Z-values of klf3+klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_22797959 | 0.88 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr18_+_5549672 | 0.75 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr1_-_44940830 | 0.63 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr2_-_35566938 | 0.62 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr12_+_34273240 | 0.56 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr3_+_55093581 | 0.54 |
ENSDART00000101709
|
hbaa2
|
hemoglobin, alpha adult 2 |
| chr24_-_40668208 | 0.48 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr24_-_40667800 | 0.46 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr15_+_28410664 | 0.43 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr20_-_54377933 | 0.42 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr11_+_37278457 | 0.38 |
ENSDART00000188946
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr21_+_27448856 | 0.37 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr19_-_677713 | 0.33 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr21_+_11244068 | 0.33 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr7_+_35229645 | 0.32 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_+_15106808 | 0.31 |
ENSDART00000141588
|
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr24_-_14212521 | 0.31 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr24_+_37461457 | 0.31 |
ENSDART00000165775
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr18_-_45617146 | 0.31 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr6_-_13022166 | 0.31 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr8_-_30204650 | 0.28 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr7_+_35229805 | 0.28 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_57235896 | 0.26 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr7_-_6356875 | 0.26 |
ENSDART00000173426
|
zgc:112234
|
zgc:112234 |
| chr25_-_28587896 | 0.25 |
ENSDART00000111908
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr4_+_54618332 | 0.23 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr1_+_44941031 | 0.23 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr5_-_24174956 | 0.22 |
ENSDART00000137631
|
si:ch211-137i24.10
|
si:ch211-137i24.10 |
| chr4_+_50401133 | 0.21 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr1_-_49434798 | 0.21 |
ENSDART00000150842
ENSDART00000150789 |
ATP5MD
|
si:dkeyp-80c12.10 |
| chr13_-_50567658 | 0.21 |
ENSDART00000132281
|
blnk
|
B cell linker |
| chr18_+_7309150 | 0.19 |
ENSDART00000052792
|
cers3b
|
ceramide synthase 3b |
| chr22_-_5051689 | 0.19 |
ENSDART00000113521
|
matk
|
megakaryocyte-associated tyrosine kinase |
| chr16_-_50918908 | 0.19 |
ENSDART00000108744
|
CU855821.1
|
|
| chr20_-_52928541 | 0.19 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr21_-_29197700 | 0.18 |
ENSDART00000172614
|
si:ch211-57b15.2
|
si:ch211-57b15.2 |
| chr25_-_36370292 | 0.18 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr7_-_6351021 | 0.18 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr19_+_19786117 | 0.17 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr21_-_280769 | 0.17 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr16_+_44768361 | 0.17 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr25_+_37348730 | 0.17 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_-_74109792 | 0.17 |
ENSDART00000174320
|
RAB21
|
zgc:154045 |
| chr6_+_120181 | 0.16 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr12_+_19030391 | 0.16 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr7_+_5968225 | 0.16 |
ENSDART00000083397
|
zgc:165555
|
zgc:165555 |
| chr20_-_52902693 | 0.15 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr20_+_48129759 | 0.15 |
ENSDART00000148494
|
tram2
|
translocation associated membrane protein 2 |
| chr15_-_5580093 | 0.15 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr17_-_24353462 | 0.14 |
ENSDART00000191021
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr16_+_23978978 | 0.14 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr23_+_9057999 | 0.14 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr3_-_1190132 | 0.14 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr25_-_207214 | 0.13 |
ENSDART00000193448
|
FP236318.3
|
|
| chr19_+_7847920 | 0.13 |
ENSDART00000132454
|
si:dkeyp-85e10.1
|
si:dkeyp-85e10.1 |
| chr10_+_15025006 | 0.13 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr7_-_2039060 | 0.13 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr3_+_60034194 | 0.13 |
ENSDART00000189858
ENSDART00000153642 |
CR352258.1
|
|
| chr12_+_34763795 | 0.12 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr1_-_59571758 | 0.12 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr21_+_21811867 | 0.11 |
ENSDART00000185752
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr21_-_35832548 | 0.11 |
ENSDART00000180840
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr8_-_34762163 | 0.11 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr11_-_8269227 | 0.11 |
ENSDART00000127202
ENSDART00000158079 ENSDART00000158748 |
si:cabz01021067.1
pimr202
|
si:cabz01021067.1 Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr8_-_3413139 | 0.11 |
ENSDART00000182673
ENSDART00000166741 ENSDART00000169430 ENSDART00000170478 |
FUT9 (1 of many)
FUT9 (1 of many)
fut9b
|
zgc:103510 zgc:165519 fucosyltransferase 9b |
| chr23_-_4930229 | 0.10 |
ENSDART00000189456
|
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr19_-_24218942 | 0.10 |
ENSDART00000189198
|
BX547993.2
|
|
| chr2_+_10007113 | 0.10 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_+_37435720 | 0.10 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr4_+_53976731 | 0.10 |
ENSDART00000165813
|
si:ch211-249c2.1
|
si:ch211-249c2.1 |
| chr6_+_46258866 | 0.09 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr6_+_41181869 | 0.09 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr7_+_4624926 | 0.09 |
ENSDART00000137387
|
si:dkey-83f18.6
|
si:dkey-83f18.6 |
| chr9_+_18576047 | 0.09 |
ENSDART00000145174
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr3_+_9332984 | 0.08 |
ENSDART00000185184
ENSDART00000183535 |
LO018550.2
|
|
| chr22_-_30948457 | 0.08 |
ENSDART00000139887
|
ssuh2.1
|
ssu-2 homolog, tandem duplicate 1 |
| chr8_-_1973220 | 0.08 |
ENSDART00000184435
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr24_+_81527 | 0.08 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr7_-_67317119 | 0.08 |
ENSDART00000183915
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr14_-_22108718 | 0.07 |
ENSDART00000054410
|
med19a
|
mediator complex subunit 19a |
| chr12_-_14143344 | 0.07 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr22_+_1006573 | 0.07 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr9_-_11551608 | 0.07 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr4_+_64562090 | 0.07 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr18_+_814328 | 0.07 |
ENSDART00000157801
ENSDART00000188492 ENSDART00000184489 |
fam219b
|
family with sequence similarity 219, member B |
| chr17_+_20569806 | 0.06 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr9_-_56420087 | 0.06 |
ENSDART00000112016
|
actr3
|
ARP3 actin related protein 3 homolog |
| chr20_+_54356272 | 0.06 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr9_-_56459660 | 0.06 |
ENSDART00000162771
|
ubxn4
|
UBX domain protein 4 |
| chr18_+_5618368 | 0.06 |
ENSDART00000159945
|
ulk3
|
unc-51 like kinase 3 |
| chr2_-_54639964 | 0.05 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr15_-_47822597 | 0.05 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr14_+_13453130 | 0.05 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr6_+_41186320 | 0.05 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr2_-_30135446 | 0.05 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr18_+_54354 | 0.04 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr23_-_1017605 | 0.04 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr3_+_1179601 | 0.04 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr11_+_45461419 | 0.03 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr23_-_1017428 | 0.03 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr2_-_58900077 | 0.02 |
ENSDART00000183362
ENSDART00000190274 |
mau2
|
MAU2 sister chromatid cohesion factor |
| chr5_-_29587351 | 0.02 |
ENSDART00000136446
ENSDART00000051434 |
entpd2a.1
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 1 |
| chr19_-_35450857 | 0.02 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr7_-_67316903 | 0.02 |
ENSDART00000159408
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr3_-_32927516 | 0.01 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr6_-_52757276 | 0.01 |
ENSDART00000159154
|
AL954861.2
|
|
| chr6_-_19665114 | 0.01 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr6_-_39344259 | 0.01 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr20_+_50052627 | 0.00 |
ENSDART00000188799
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf3+klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.0 | 0.3 | GO:0048246 | organ regeneration(GO:0031100) macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.6 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |