Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
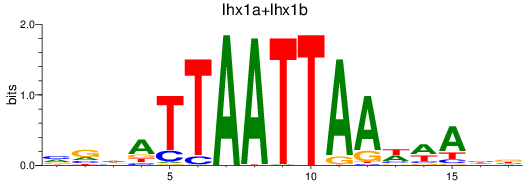
Results for lhx1a+lhx1b
Z-value: 0.57
Transcription factors associated with lhx1a+lhx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx1b
|
ENSDARG00000007944 | LIM homeobox 1b |
|
lhx1a
|
ENSDARG00000014018 | LIM homeobox 1a |
|
lhx1b
|
ENSDARG00000111635 | LIM homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx1b | dr11_v1_chr5_+_56268436_56268436 | 0.21 | 7.4e-01 | Click! |
| lhx1a | dr11_v1_chr15_-_27710513_27710575 | 0.20 | 7.4e-01 | Click! |
Activity profile of lhx1a+lhx1b motif
Sorted Z-values of lhx1a+lhx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43297546 | 0.37 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr15_-_43284021 | 0.28 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr9_-_9415000 | 0.26 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr20_+_11731039 | 0.25 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr3_-_19368435 | 0.24 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr1_+_51039558 | 0.23 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr22_-_19552796 | 0.22 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr7_-_38658411 | 0.22 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr8_+_50953776 | 0.21 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr24_+_2495197 | 0.20 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr10_-_11261565 | 0.20 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr23_+_4689626 | 0.19 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr19_+_2631565 | 0.19 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_+_10318089 | 0.18 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr19_+_46158078 | 0.18 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr23_-_16485190 | 0.18 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr4_+_12966640 | 0.17 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr4_+_16885854 | 0.16 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr2_-_16217344 | 0.15 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_9669717 | 0.15 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr3_-_15999501 | 0.15 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr21_-_20939488 | 0.15 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr3_-_32818607 | 0.14 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_+_4054704 | 0.14 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr3_+_26342768 | 0.13 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr8_-_49728590 | 0.13 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr12_+_45200744 | 0.13 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr9_-_27398369 | 0.13 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr16_+_54209504 | 0.13 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr19_-_5769728 | 0.12 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr6_-_35052145 | 0.12 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr19_-_5769553 | 0.11 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr1_-_51038885 | 0.11 |
ENSDART00000035150
|
spast
|
spastin |
| chr10_+_11261576 | 0.11 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr23_-_31969786 | 0.11 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr5_-_31901468 | 0.11 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr25_-_13490744 | 0.11 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr3_-_29941357 | 0.10 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr21_-_2415808 | 0.10 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr13_+_11440389 | 0.10 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr6_-_50730749 | 0.10 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr24_+_26328787 | 0.10 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr15_-_20939579 | 0.10 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr22_-_28226948 | 0.10 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr1_+_6172786 | 0.10 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr22_-_22242884 | 0.10 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr5_+_61301525 | 0.10 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr2_+_38373272 | 0.10 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr4_+_77943184 | 0.10 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr23_-_4409668 | 0.10 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr17_-_16422654 | 0.10 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr6_+_11989537 | 0.09 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr8_+_19504011 | 0.09 |
ENSDART00000158859
ENSDART00000159044 |
sec22bb
|
SEC22 homolog B, vesicle trafficking protein b |
| chr23_-_17509656 | 0.09 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr25_-_21031007 | 0.09 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr22_-_12160283 | 0.09 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr2_+_105748 | 0.09 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr6_-_40029423 | 0.09 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr20_-_28642061 | 0.08 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr11_-_40204621 | 0.08 |
ENSDART00000086287
|
znf362a
|
zinc finger protein 362a |
| chr24_-_26328721 | 0.08 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr5_+_41477954 | 0.08 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr13_+_33268657 | 0.08 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr4_+_5255041 | 0.08 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr6_+_35052721 | 0.08 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr1_-_669717 | 0.08 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr22_-_10156581 | 0.08 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr23_+_20640484 | 0.07 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr9_-_14504834 | 0.07 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr1_-_45616470 | 0.07 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_-_30839763 | 0.07 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr23_-_16484383 | 0.07 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr20_-_45812144 | 0.07 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr5_+_60590796 | 0.06 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_-_35649000 | 0.06 |
ENSDART00000053310
|
tmem18
|
transmembrane protein 18 |
| chr10_-_11261386 | 0.06 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr3_-_26806032 | 0.06 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr6_-_55399214 | 0.06 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr10_+_17235370 | 0.06 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr17_+_45405821 | 0.06 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr16_+_13965923 | 0.05 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr3_-_26787430 | 0.05 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr5_+_41477526 | 0.05 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr1_+_54124209 | 0.05 |
ENSDART00000187730
|
LO017722.1
|
|
| chr23_-_18567088 | 0.05 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr3_+_36424055 | 0.05 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr2_-_30668580 | 0.05 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr16_+_2820340 | 0.05 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr8_+_7801060 | 0.05 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr6_+_7444899 | 0.04 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr8_-_53926228 | 0.04 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr6_-_12912606 | 0.04 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr24_+_28953089 | 0.04 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_+_21731382 | 0.04 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr16_-_28856112 | 0.04 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr25_-_25058508 | 0.04 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr12_+_48480632 | 0.04 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr10_+_45089820 | 0.04 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr4_+_72723304 | 0.04 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr21_+_25802190 | 0.03 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr23_+_4709607 | 0.03 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr16_-_12173399 | 0.03 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr7_+_22809905 | 0.03 |
ENSDART00000166900
ENSDART00000143455 ENSDART00000126037 |
sf1
|
splicing factor 1 |
| chr19_+_7424347 | 0.03 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr24_-_35767501 | 0.03 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr17_-_200316 | 0.03 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr25_+_4751879 | 0.03 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr6_-_11768198 | 0.03 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr11_-_25733910 | 0.03 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr25_+_4855549 | 0.03 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr16_-_42965192 | 0.03 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr25_+_18711804 | 0.03 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr24_+_26329018 | 0.02 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr9_-_47472998 | 0.02 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr6_-_35052388 | 0.02 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr6_-_34838397 | 0.02 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr8_-_12867128 | 0.02 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr23_+_17509794 | 0.02 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr9_+_34641237 | 0.02 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr24_-_40860603 | 0.02 |
ENSDART00000188032
|
CU633479.7
|
|
| chr7_+_4474880 | 0.02 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr4_-_1801519 | 0.02 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr21_-_13972745 | 0.02 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr21_-_35419486 | 0.02 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr19_+_5480327 | 0.02 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr3_+_39853788 | 0.01 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr10_+_42423318 | 0.01 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr6_+_39905021 | 0.01 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr6_-_44402358 | 0.01 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr9_+_24159725 | 0.01 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr9_+_24159280 | 0.01 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr12_+_7497882 | 0.01 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr10_+_16584382 | 0.01 |
ENSDART00000112039
|
CR790388.1
|
|
| chr5_-_30620625 | 0.01 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr9_+_54039006 | 0.01 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr15_-_35112937 | 0.01 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr16_+_23303859 | 0.00 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr21_-_17482465 | 0.00 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr17_+_28533102 | 0.00 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx1a+lhx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |