Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
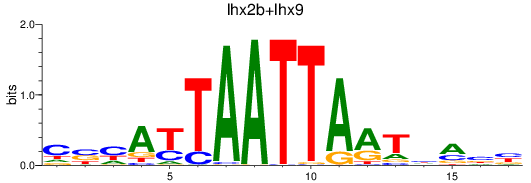
Results for lhx2b+lhx9
Z-value: 0.46
Transcription factors associated with lhx2b+lhx9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx2b
|
ENSDARG00000031222 | LIM homeobox 2b |
|
lhx9
|
ENSDARG00000056979 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx9 | dr11_v1_chr22_-_23253481_23253481 | -0.67 | 2.2e-01 | Click! |
| lhx2b | dr11_v1_chr8_+_3085120_3085219 | -0.57 | 3.2e-01 | Click! |
Activity profile of lhx2b+lhx9 motif
Sorted Z-values of lhx2b+lhx9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_63302944 | 0.47 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr25_-_21031007 | 0.46 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr9_-_9415000 | 0.38 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr12_+_37401331 | 0.37 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr21_+_25236297 | 0.36 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr14_+_901847 | 0.29 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr23_-_33350990 | 0.28 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr19_+_2631565 | 0.28 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr8_-_25771474 | 0.28 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr10_+_2742499 | 0.28 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr19_+_43297546 | 0.28 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr20_+_28364742 | 0.27 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr2_-_36918709 | 0.27 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr12_-_4243268 | 0.27 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr21_+_25777425 | 0.26 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr3_+_13559199 | 0.26 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr14_-_4145594 | 0.25 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr6_-_35046735 | 0.25 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr23_+_36460239 | 0.24 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr12_-_35830625 | 0.24 |
ENSDART00000180028
|
CU459056.1
|
|
| chr8_+_34731982 | 0.23 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr1_+_9966384 | 0.22 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr6_+_39905021 | 0.22 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr22_-_10156581 | 0.20 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr19_+_7895086 | 0.20 |
ENSDART00000132180
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr20_-_35750810 | 0.20 |
ENSDART00000153072
|
adgrf8
|
adhesion G protein-coupled receptor F8 |
| chr9_+_21165484 | 0.20 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr8_-_20230559 | 0.19 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr7_-_54320088 | 0.19 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr13_+_7442023 | 0.19 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr10_+_17776981 | 0.19 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr14_+_15257658 | 0.19 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr11_+_705727 | 0.19 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr19_+_43780970 | 0.19 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr3_+_29941777 | 0.19 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr25_+_3217419 | 0.19 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr4_+_77943184 | 0.18 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr23_-_16485190 | 0.18 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr22_+_5687615 | 0.18 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr4_-_71904377 | 0.18 |
ENSDART00000191852
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr22_+_19366866 | 0.17 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr10_-_34002185 | 0.17 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr5_+_27897504 | 0.17 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr4_-_9891874 | 0.16 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr4_+_53119675 | 0.16 |
ENSDART00000167470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr22_+_5532003 | 0.16 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr21_-_44081540 | 0.16 |
ENSDART00000130833
|
FO704810.1
|
|
| chr5_-_30620625 | 0.16 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr9_-_30555725 | 0.16 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr12_-_18578432 | 0.16 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr6_-_43677125 | 0.16 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr23_+_4709607 | 0.16 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr25_+_8356707 | 0.15 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr17_-_27048537 | 0.15 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr5_-_30715225 | 0.15 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr5_-_57723929 | 0.15 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr20_+_25586099 | 0.15 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr5_+_27898226 | 0.15 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr3_-_59297532 | 0.15 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr2_+_36112273 | 0.14 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr23_-_36446307 | 0.14 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr24_-_26622423 | 0.14 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr10_+_36650222 | 0.14 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr10_+_29850330 | 0.13 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr8_-_21091961 | 0.13 |
ENSDART00000100281
|
aldh9a1a.2
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 2 |
| chr22_+_508290 | 0.13 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr5_+_2815021 | 0.13 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr25_+_11008419 | 0.13 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr3_-_50443607 | 0.13 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr3_+_26145013 | 0.13 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr18_-_8877077 | 0.13 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr4_-_42408339 | 0.12 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr22_+_16535575 | 0.12 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr17_+_8799661 | 0.12 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_+_32430982 | 0.12 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr12_+_20641102 | 0.12 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr25_+_29474982 | 0.12 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr12_+_20641471 | 0.12 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr20_-_43663494 | 0.12 |
ENSDART00000144564
|
BX470188.1
|
|
| chr14_-_7207961 | 0.12 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr14_-_14659023 | 0.12 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr23_-_1660708 | 0.12 |
ENSDART00000175138
|
CU693481.1
|
|
| chr2_+_9990491 | 0.12 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr8_-_20230802 | 0.12 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr2_+_105748 | 0.12 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr4_+_966061 | 0.12 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr9_-_43644261 | 0.12 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr7_-_50410524 | 0.12 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr23_-_29394505 | 0.11 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr3_-_32079916 | 0.11 |
ENSDART00000040900
|
baxb
|
BCL2 associated X, apoptosis regulator b |
| chr24_-_32582880 | 0.11 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr15_-_43284021 | 0.11 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_-_39969988 | 0.11 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr21_-_26114886 | 0.11 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr23_-_30041065 | 0.11 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr23_-_36303216 | 0.11 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr8_+_11425048 | 0.11 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr25_+_22320738 | 0.11 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_+_17167876 | 0.10 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr6_+_612594 | 0.10 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr17_-_23616626 | 0.10 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr2_-_10877765 | 0.10 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr23_+_384850 | 0.10 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr3_-_29941357 | 0.10 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr1_+_44127292 | 0.10 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr8_-_30204650 | 0.10 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr16_-_11798994 | 0.10 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr17_+_8799451 | 0.10 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr12_-_18578218 | 0.10 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr13_-_31008275 | 0.10 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr5_+_13647288 | 0.09 |
ENSDART00000099660
ENSDART00000139199 |
h2afva
|
H2A histone family, member Va |
| chr20_+_25225112 | 0.09 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr19_+_12237945 | 0.09 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr7_-_3836896 | 0.09 |
ENSDART00000136227
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr7_-_3836635 | 0.09 |
ENSDART00000104350
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr10_+_11261576 | 0.09 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr2_+_48073972 | 0.09 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr15_-_38129845 | 0.09 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr6_-_39270851 | 0.09 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr2_-_9989919 | 0.09 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr3_-_31079186 | 0.09 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr1_+_51721851 | 0.09 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr1_-_55118745 | 0.09 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr22_+_7738966 | 0.09 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr8_+_28695914 | 0.09 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr4_-_62714083 | 0.09 |
ENSDART00000188299
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr20_+_48116476 | 0.09 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr21_-_1644414 | 0.08 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr17_-_20558961 | 0.08 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr3_-_10739625 | 0.08 |
ENSDART00000156144
|
zgc:112965
|
zgc:112965 |
| chr7_+_71664624 | 0.08 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr23_+_11347313 | 0.08 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr3_-_15999501 | 0.08 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr9_+_23895711 | 0.08 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr5_+_38752287 | 0.08 |
ENSDART00000133571
|
cxcl11.8
|
chemokine (C-X-C motif) ligand 11, duplicate 8 |
| chr3_-_53114299 | 0.08 |
ENSDART00000109390
|
AL954361.1
|
|
| chr2_+_20406399 | 0.08 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr5_-_11809710 | 0.08 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr23_-_42232124 | 0.08 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr24_+_2495197 | 0.08 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr11_+_42474694 | 0.08 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr3_-_57666518 | 0.08 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr3_-_32818607 | 0.07 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_-_66823750 | 0.07 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr19_+_14573998 | 0.07 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr22_+_37631234 | 0.07 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr1_+_47499888 | 0.07 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr6_+_8598428 | 0.07 |
ENSDART00000032118
|
ube2g2
|
ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) |
| chr22_-_9183944 | 0.07 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr22_+_737211 | 0.07 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr6_+_12527725 | 0.07 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr16_+_54209504 | 0.07 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr19_+_43359075 | 0.07 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr10_+_44373349 | 0.07 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr13_+_33268657 | 0.07 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr19_-_41518922 | 0.07 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr13_-_42560662 | 0.07 |
ENSDART00000124898
|
CR792417.1
|
|
| chr15_-_40157165 | 0.07 |
ENSDART00000192991
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr10_-_31015535 | 0.07 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr7_-_8738827 | 0.07 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr4_-_67980261 | 0.07 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr2_-_7845110 | 0.07 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr15_-_40157331 | 0.06 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr25_+_28825657 | 0.06 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr12_-_18577983 | 0.06 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr19_+_1688727 | 0.06 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr25_-_18739924 | 0.06 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr7_-_67248829 | 0.06 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr16_+_24733741 | 0.06 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr4_+_5255041 | 0.06 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr20_+_5985329 | 0.06 |
ENSDART00000165489
|
cep128
|
centrosomal protein 128 |
| chr16_+_28994709 | 0.06 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr7_+_57088920 | 0.06 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_+_44571200 | 0.06 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr12_-_28363111 | 0.06 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr13_+_18321140 | 0.06 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr21_+_11885404 | 0.06 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr20_-_49889111 | 0.06 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr23_+_16815353 | 0.06 |
ENSDART00000142789
ENSDART00000143424 |
zgc:100832
|
zgc:100832 |
| chr9_+_37152564 | 0.06 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr3_+_23029934 | 0.05 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr8_+_45334255 | 0.05 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_-_51355465 | 0.05 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr12_+_20587179 | 0.05 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr15_-_40157513 | 0.05 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr15_+_21262917 | 0.05 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr10_-_42923385 | 0.05 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr8_-_12867128 | 0.05 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr8_+_28259347 | 0.05 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr23_-_18567088 | 0.05 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr1_-_55248496 | 0.05 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr24_-_7995960 | 0.05 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr2_-_7843451 | 0.05 |
ENSDART00000163265
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr22_+_37631034 | 0.05 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr5_+_1624359 | 0.05 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr8_+_52637507 | 0.05 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr22_+_1708760 | 0.05 |
ENSDART00000170471
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr10_-_43771447 | 0.05 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr5_-_23843636 | 0.05 |
ENSDART00000193280
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr14_+_46313396 | 0.05 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx2b+lhx9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |