Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
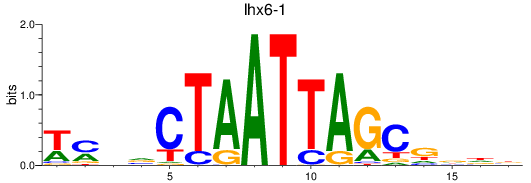
Results for lhx6-1
Z-value: 0.80
Transcription factors associated with lhx6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6-1
|
ENSDARG00000052165 | si_ch211-236k19.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-236k19.2 | dr11_v1_chr5_-_64900552_64900552 | -0.24 | 7.0e-01 | Click! |
Activity profile of lhx6-1 motif
Sorted Z-values of lhx6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_6751405 | 0.59 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr21_+_6751760 | 0.47 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr25_+_3327071 | 0.46 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr25_+_3326885 | 0.41 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr13_-_4707018 | 0.34 |
ENSDART00000128422
|
oit3
|
oncoprotein induced transcript 3 |
| chr8_-_45277370 | 0.33 |
ENSDART00000146364
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr2_+_6255434 | 0.31 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr16_-_28658341 | 0.30 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr10_+_7709724 | 0.29 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr19_+_9295244 | 0.27 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr22_-_22719440 | 0.27 |
ENSDART00000166794
|
nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_30392613 | 0.26 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr3_-_26017831 | 0.25 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_23407720 | 0.23 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr15_+_45563656 | 0.22 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr2_-_38284648 | 0.22 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr2_+_6253246 | 0.22 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr12_+_47446158 | 0.22 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr11_+_5588122 | 0.22 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr23_-_41651759 | 0.22 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr16_+_23975930 | 0.21 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr1_+_29068654 | 0.21 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr9_+_44994214 | 0.21 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr17_+_30704068 | 0.20 |
ENSDART00000062793
|
apoba
|
apolipoprotein Ba |
| chr24_-_9979342 | 0.20 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr19_-_5805923 | 0.19 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr4_+_11375894 | 0.19 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr16_+_23976227 | 0.18 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr15_+_44366556 | 0.18 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr2_+_50608099 | 0.17 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_-_9882821 | 0.17 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr12_-_22238004 | 0.17 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr10_+_34001444 | 0.17 |
ENSDART00000149934
|
kl
|
klotho |
| chr23_-_30787932 | 0.17 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr6_+_57541776 | 0.17 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr13_+_771403 | 0.17 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr9_+_219124 | 0.16 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr16_+_47207691 | 0.16 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr5_+_22098591 | 0.16 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr14_-_14659023 | 0.16 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr10_-_34915886 | 0.15 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr6_+_27151940 | 0.15 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr3_+_46559639 | 0.15 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr17_-_37214196 | 0.15 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr3_+_33345348 | 0.15 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr25_+_21324588 | 0.15 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr24_-_33366188 | 0.14 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr6_+_25261297 | 0.14 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr20_+_23947004 | 0.14 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr13_-_25198025 | 0.14 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr13_+_8954667 | 0.13 |
ENSDART00000100241
|
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_+_16188761 | 0.13 |
ENSDART00000193244
|
ctxn3
|
cortexin 3 |
| chr12_+_1469090 | 0.13 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr18_+_38908903 | 0.13 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr6_+_34511886 | 0.13 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_+_19708508 | 0.13 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr7_+_69528850 | 0.12 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr4_-_28158335 | 0.12 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr16_+_39159752 | 0.12 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_-_25004553 | 0.12 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr23_+_28582865 | 0.12 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr25_+_13498188 | 0.12 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr5_-_62306819 | 0.12 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr18_+_21408794 | 0.12 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_-_25275364 | 0.12 |
ENSDART00000163782
ENSDART00000145420 ENSDART00000133718 ENSDART00000055492 |
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr22_-_16180467 | 0.12 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr19_-_5103313 | 0.11 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_-_67629263 | 0.11 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_6519691 | 0.11 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr10_+_15777064 | 0.11 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_+_56937548 | 0.11 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr15_-_22074315 | 0.11 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr1_-_46981134 | 0.11 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_+_4646194 | 0.11 |
ENSDART00000092344
|
LO017700.1
|
|
| chr25_-_1323623 | 0.11 |
ENSDART00000156532
ENSDART00000157163 ENSDART00000156062 ENSDART00000082447 ENSDART00000189175 |
calml4b
|
calmodulin-like 4b |
| chr15_-_9593532 | 0.11 |
ENSDART00000169912
|
gab2
|
GRB2-associated binding protein 2 |
| chr4_+_9467049 | 0.11 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr1_+_30723677 | 0.11 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr17_-_200316 | 0.11 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr18_+_11506561 | 0.11 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr6_+_40951227 | 0.11 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr2_-_3038904 | 0.11 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr25_+_26921480 | 0.11 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr22_-_20011476 | 0.11 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr8_+_49975160 | 0.11 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr5_-_12407194 | 0.10 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr21_-_39177564 | 0.10 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr11_+_38280454 | 0.10 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr21_+_43404945 | 0.10 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr22_-_7050 | 0.10 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr5_+_64842730 | 0.10 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr22_-_29083070 | 0.10 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr7_+_30787903 | 0.10 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr5_+_20257225 | 0.10 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr16_-_29437373 | 0.10 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr9_-_12886108 | 0.10 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr10_+_44584614 | 0.10 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr2_+_56168619 | 0.10 |
ENSDART00000021011
ENSDART00000180271 |
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr19_+_5604241 | 0.09 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr17_+_1063988 | 0.09 |
ENSDART00000160400
|
gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr12_-_42214 | 0.09 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr3_-_46817499 | 0.09 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr11_-_41130239 | 0.09 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr24_-_18809433 | 0.09 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr4_+_77957611 | 0.09 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr14_-_47815467 | 0.09 |
ENSDART00000166827
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr16_-_40841363 | 0.09 |
ENSDART00000163380
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr9_-_10460087 | 0.09 |
ENSDART00000186001
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr3_-_30625219 | 0.09 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr3_-_23406964 | 0.09 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr2_-_15031858 | 0.09 |
ENSDART00000191478
|
hccsa.1
|
holocytochrome c synthase a |
| chr10_-_34916208 | 0.09 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr5_+_42388464 | 0.09 |
ENSDART00000191596
|
BX548073.13
|
|
| chr1_+_33668236 | 0.09 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr8_-_23780334 | 0.09 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr17_+_8212477 | 0.09 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr4_-_9191220 | 0.09 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr21_+_26390549 | 0.08 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr5_-_27993972 | 0.08 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr5_-_64831207 | 0.08 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr8_-_16464453 | 0.08 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr13_+_2442841 | 0.08 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr11_-_15296805 | 0.08 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr7_+_49272702 | 0.08 |
ENSDART00000083389
|
abtb2a
|
ankyrin repeat and BTB (POZ) domain containing 2a |
| chr9_+_17983463 | 0.08 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr4_-_23873924 | 0.08 |
ENSDART00000100391
|
usp6nl
|
USP6 N-terminal like |
| chr22_-_21897203 | 0.08 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr8_-_39822917 | 0.08 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr20_-_47188966 | 0.08 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr11_+_45323455 | 0.08 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr19_+_20274264 | 0.08 |
ENSDART00000183513
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr12_-_3940768 | 0.08 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr19_-_20430892 | 0.08 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr7_+_54642005 | 0.08 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr24_-_31843173 | 0.08 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr13_+_24584401 | 0.08 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr4_+_21129752 | 0.08 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_+_20274491 | 0.07 |
ENSDART00000090860
ENSDART00000151693 |
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr3_-_60827402 | 0.07 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr4_-_29753895 | 0.07 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr11_-_16021424 | 0.07 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr4_+_11384891 | 0.07 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr5_+_25733774 | 0.07 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr1_+_30723380 | 0.07 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr7_+_7151832 | 0.07 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr25_+_459901 | 0.07 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr24_-_24724233 | 0.07 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr4_-_73756673 | 0.07 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr5_+_42381273 | 0.07 |
ENSDART00000142387
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr19_-_1871415 | 0.07 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr5_-_25733745 | 0.07 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr18_-_7112882 | 0.07 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr23_-_33038423 | 0.07 |
ENSDART00000180539
|
plxna2
|
plexin A2 |
| chr8_+_8947623 | 0.07 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr23_-_4225606 | 0.06 |
ENSDART00000014152
ENSDART00000133341 |
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr3_+_32365811 | 0.06 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr20_-_29532939 | 0.06 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr1_-_29139141 | 0.06 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_-_105100 | 0.06 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr8_-_15129573 | 0.06 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr20_-_13774826 | 0.06 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr16_-_25568512 | 0.06 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr19_-_46088429 | 0.06 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr24_+_9178064 | 0.06 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr13_-_10730056 | 0.06 |
ENSDART00000126725
|
ppm1ba
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ba |
| chr21_-_40398932 | 0.06 |
ENSDART00000162500
|
nup98
|
nucleoporin 98 |
| chr6_-_40922971 | 0.06 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr22_-_33679277 | 0.06 |
ENSDART00000169948
|
FO904977.1
|
|
| chr17_-_37195354 | 0.06 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr2_-_55298075 | 0.05 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr17_-_37195163 | 0.05 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr2_-_21621878 | 0.05 |
ENSDART00000189540
|
zgc:55781
|
zgc:55781 |
| chr24_+_1023839 | 0.05 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr4_+_9400012 | 0.05 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr11_+_2855430 | 0.05 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr14_-_12020653 | 0.05 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr12_+_36428052 | 0.05 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr2_-_40890004 | 0.05 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr7_+_42460302 | 0.05 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr19_+_1673599 | 0.05 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr3_-_13147310 | 0.05 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr12_+_1469327 | 0.05 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr23_-_14766902 | 0.05 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr17_+_10593398 | 0.05 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr7_+_51795667 | 0.05 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr24_+_17260329 | 0.05 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr10_+_16036246 | 0.05 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr5_+_28308774 | 0.05 |
ENSDART00000087708
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr19_+_9231766 | 0.05 |
ENSDART00000190617
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr24_+_17260001 | 0.05 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr8_+_14855471 | 0.05 |
ENSDART00000134775
|
si:dkey-21p1.3
|
si:dkey-21p1.3 |
| chr6_+_13779857 | 0.05 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr7_+_15872357 | 0.05 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr5_+_13394543 | 0.05 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr18_+_1145571 | 0.05 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr22_-_621609 | 0.05 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr11_-_44876005 | 0.05 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr3_+_726000 | 0.05 |
ENSDART00000158510
|
dicp1.17
|
diverse immunoglobulin domain-containing protein 1.17 |
| chr16_+_35379720 | 0.05 |
ENSDART00000170497
|
si:dkey-34d22.2
|
si:dkey-34d22.2 |
| chr6_-_47840465 | 0.04 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |