Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
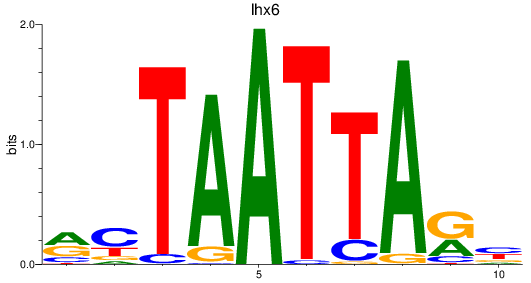
Results for lhx6
Z-value: 0.34
Transcription factors associated with lhx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6
|
ENSDARG00000006896 | LIM homeobox 6 |
|
lhx6
|
ENSDARG00000112520 | LIM homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx6 | dr11_v1_chr10_+_9372702_9372702 | 0.53 | 3.6e-01 | Click! |
Activity profile of lhx6 motif
Sorted Z-values of lhx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_64732270 | 0.33 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr17_+_15433518 | 0.30 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 0.29 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_+_64732036 | 0.29 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr4_+_21129752 | 0.23 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr5_+_22098591 | 0.23 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr7_+_30787903 | 0.22 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr8_+_16025554 | 0.22 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_50608099 | 0.21 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_+_233143 | 0.20 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr11_-_42554290 | 0.20 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr3_-_46811611 | 0.19 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_-_33872092 | 0.19 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr7_-_49594995 | 0.18 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr6_+_39232245 | 0.18 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr6_-_55585423 | 0.17 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr11_+_38280454 | 0.17 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr5_+_36768674 | 0.16 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr17_-_200316 | 0.16 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr5_-_50992690 | 0.16 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr10_-_27049170 | 0.16 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr18_+_8320165 | 0.16 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr16_+_34531486 | 0.16 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr8_+_49778486 | 0.16 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr23_+_40460333 | 0.16 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr14_-_33872616 | 0.15 |
ENSDART00000162840
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr16_+_46111849 | 0.15 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr13_+_38521152 | 0.15 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr2_+_6253246 | 0.15 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr2_+_20332044 | 0.14 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr17_-_36896560 | 0.14 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr19_-_9882821 | 0.14 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr25_-_13842618 | 0.14 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr4_+_4849789 | 0.13 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr3_-_46817499 | 0.13 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr1_+_25801648 | 0.13 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr9_+_17983463 | 0.13 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr20_-_29864390 | 0.13 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr10_+_42423318 | 0.13 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr9_-_20372977 | 0.13 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr16_+_47207691 | 0.13 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr9_+_30108641 | 0.13 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr8_+_49778756 | 0.13 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr23_+_28582865 | 0.13 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr3_-_23406964 | 0.12 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr9_-_43538328 | 0.12 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr22_+_27090136 | 0.12 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr4_+_11384891 | 0.12 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr14_-_30587814 | 0.12 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr15_-_33925851 | 0.12 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr13_-_31435137 | 0.12 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr4_-_1801519 | 0.12 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr11_+_18873619 | 0.12 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr2_+_40294313 | 0.11 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr2_-_32558795 | 0.11 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr5_-_64831207 | 0.11 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr14_+_14662116 | 0.11 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr6_-_12172424 | 0.11 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr3_+_34919810 | 0.11 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr12_-_4781801 | 0.11 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr1_-_44701313 | 0.11 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr13_+_15838151 | 0.11 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr6_-_6487876 | 0.10 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr17_-_40956035 | 0.10 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr5_-_16996482 | 0.10 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr7_-_28148310 | 0.10 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr14_-_33105434 | 0.10 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr21_+_32820175 | 0.10 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr6_-_51386656 | 0.10 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr4_-_28158335 | 0.10 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr10_+_21726853 | 0.10 |
ENSDART00000162869
|
pcdh1g14
|
protocadherin 1 gamma 14 |
| chr7_+_56577522 | 0.09 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr19_-_30524952 | 0.09 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr10_+_21867307 | 0.09 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr11_+_28476298 | 0.09 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr22_-_24818066 | 0.09 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr13_-_29420885 | 0.09 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr24_+_19591893 | 0.09 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr17_+_3379673 | 0.09 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr3_+_62353650 | 0.09 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr3_+_17537352 | 0.09 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr14_+_25817628 | 0.08 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr16_+_23975930 | 0.08 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr9_-_37748513 | 0.08 |
ENSDART00000188967
ENSDART00000187886 |
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr8_+_7756893 | 0.08 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr7_+_56577906 | 0.08 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr14_+_8940326 | 0.08 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr19_+_10661520 | 0.08 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr21_-_14175838 | 0.08 |
ENSDART00000111659
|
whrna
|
whirlin a |
| chr5_-_58112032 | 0.08 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr23_-_6660985 | 0.08 |
ENSDART00000162405
|
CR450824.2
|
|
| chr25_-_19224298 | 0.08 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr11_+_41540862 | 0.08 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr11_+_18873113 | 0.08 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr15_-_22074315 | 0.08 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr11_-_39044595 | 0.08 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr21_+_26748141 | 0.08 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr24_+_16547035 | 0.08 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr5_+_37903790 | 0.08 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr1_+_18811679 | 0.08 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr21_+_42226113 | 0.08 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr23_-_46020226 | 0.08 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr24_-_35767501 | 0.08 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr21_+_28958471 | 0.08 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_+_45918981 | 0.07 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr9_+_34641237 | 0.07 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr13_+_2442841 | 0.07 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr1_-_50859053 | 0.07 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr20_+_41021054 | 0.07 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr17_-_43594864 | 0.07 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr5_+_61301525 | 0.07 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr17_-_47220934 | 0.07 |
ENSDART00000158740
|
alk
|
ALK receptor tyrosine kinase |
| chr20_-_46362606 | 0.07 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr9_-_51436377 | 0.07 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr9_-_37749973 | 0.07 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr15_-_9272328 | 0.07 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_+_56645710 | 0.07 |
ENSDART00000193978
|
CR759836.1
|
|
| chr4_-_73756673 | 0.07 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr5_+_19094153 | 0.07 |
ENSDART00000186525
ENSDART00000064752 |
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr19_-_5103313 | 0.06 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_+_11267829 | 0.06 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr5_-_41307550 | 0.06 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr2_-_30668580 | 0.06 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr24_+_35881124 | 0.06 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr6_+_21001264 | 0.06 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr5_-_63515210 | 0.06 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr17_-_8570257 | 0.06 |
ENSDART00000154713
ENSDART00000121488 |
fzd3b
|
frizzled class receptor 3b |
| chr1_-_15797663 | 0.06 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr18_-_2433011 | 0.06 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr6_+_9289802 | 0.06 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr12_-_35393211 | 0.06 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr16_-_26296477 | 0.06 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr6_-_12275836 | 0.06 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr11_+_5588122 | 0.06 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr4_-_17629444 | 0.05 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr18_+_17827149 | 0.05 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr3_-_60175470 | 0.05 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr16_+_23976227 | 0.05 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr24_-_9979342 | 0.05 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr24_-_25004553 | 0.05 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr5_-_19006290 | 0.05 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr23_+_27782071 | 0.05 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr5_+_19094462 | 0.05 |
ENSDART00000190596
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr9_-_35633827 | 0.05 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr7_-_31759602 | 0.05 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr24_-_21923930 | 0.05 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr17_+_26352372 | 0.05 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr1_-_12109216 | 0.05 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr1_-_46981134 | 0.05 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr4_+_12612723 | 0.05 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr9_+_25776971 | 0.05 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr4_+_9400012 | 0.05 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr15_-_14552101 | 0.05 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr23_-_10786400 | 0.05 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr4_+_12612145 | 0.05 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr1_+_33668236 | 0.05 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr13_+_19322686 | 0.05 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr19_+_22062202 | 0.05 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr20_-_16171297 | 0.05 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr11_-_40257225 | 0.05 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr15_+_17345609 | 0.05 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr7_-_12464412 | 0.04 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr3_-_56541723 | 0.04 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr18_+_7639401 | 0.04 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr22_-_8725768 | 0.04 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr5_-_51619742 | 0.04 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr12_-_14143344 | 0.04 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr24_+_13316737 | 0.04 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr21_-_39177564 | 0.04 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr1_+_13930625 | 0.04 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr5_+_29851433 | 0.04 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr11_-_40728380 | 0.04 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr21_-_32060993 | 0.04 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr9_-_746317 | 0.04 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr8_+_29986265 | 0.04 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr1_-_29139141 | 0.04 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_+_31222433 | 0.04 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr10_-_5847655 | 0.04 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr2_+_26237322 | 0.04 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr20_-_23426339 | 0.04 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_10593398 | 0.03 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr14_+_46274611 | 0.03 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr5_-_67241633 | 0.03 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr7_-_7692992 | 0.03 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr16_-_42056137 | 0.03 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr19_-_5103141 | 0.03 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr2_+_37140448 | 0.03 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr7_-_31759394 | 0.03 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr9_+_38372216 | 0.03 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr24_+_35787629 | 0.03 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr4_-_5652030 | 0.03 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr7_-_7692723 | 0.03 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr24_+_40860320 | 0.03 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr1_+_51191049 | 0.03 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr20_-_40755614 | 0.03 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr3_-_43356082 | 0.03 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr20_-_1141722 | 0.03 |
ENSDART00000152675
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr17_+_24722646 | 0.03 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr18_+_27738349 | 0.03 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr2_+_35728033 | 0.03 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
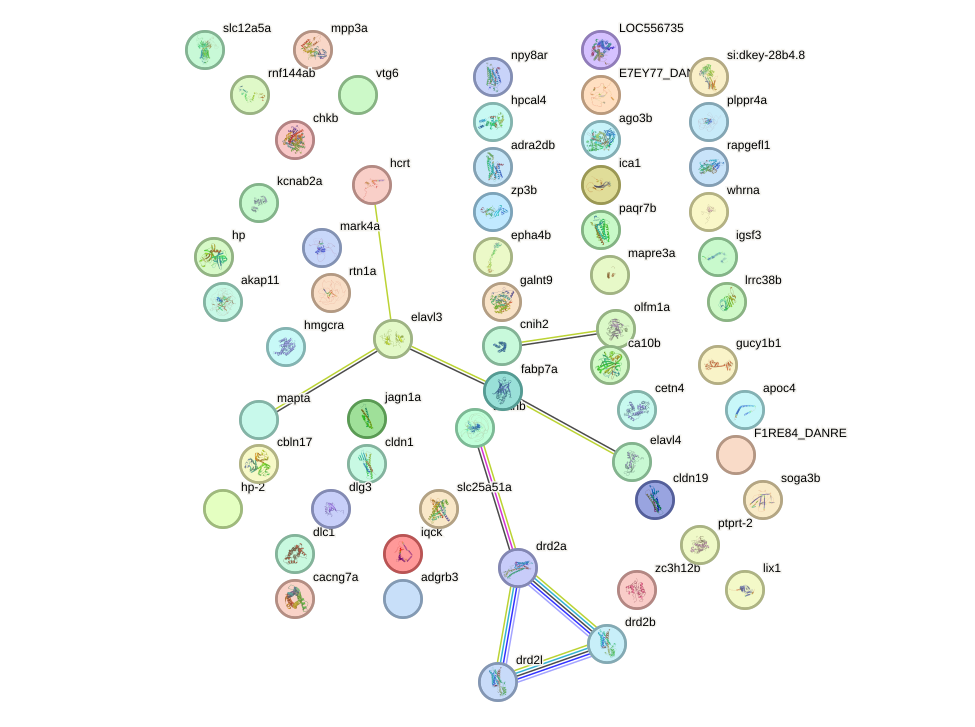
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.6 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0051481 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) cell-cell recognition(GO:0009988) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |