Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
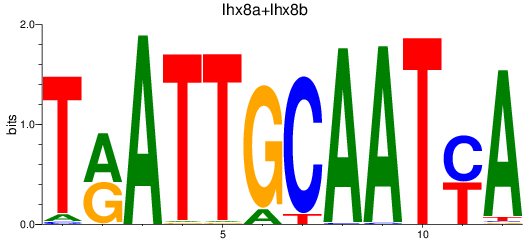
Results for lhx8a+lhx8b
Z-value: 0.69
Transcription factors associated with lhx8a+lhx8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx8a
|
ENSDARG00000002330 | LIM homeobox 8a |
|
lhx8b
|
ENSDARG00000042145 | LIM homeobox 8b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx8b | dr11_v1_chr8_-_17926814_17926814 | 0.16 | 7.9e-01 | Click! |
| lhx8a | dr11_v1_chr2_+_11206317_11206317 | -0.06 | 9.2e-01 | Click! |
Activity profile of lhx8a+lhx8b motif
Sorted Z-values of lhx8a+lhx8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_7546259 | 0.53 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr5_+_32247310 | 0.47 |
ENSDART00000182649
|
myha
|
myosin, heavy chain a |
| chr10_-_43568239 | 0.44 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr15_-_23645810 | 0.40 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr8_-_53198154 | 0.38 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr17_-_6730247 | 0.37 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr19_-_31042570 | 0.36 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr8_+_3820134 | 0.36 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr24_+_792429 | 0.35 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr14_+_17197132 | 0.33 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr12_-_4756478 | 0.33 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr18_+_924949 | 0.31 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr9_-_41277347 | 0.31 |
ENSDART00000181213
|
glsb
|
glutaminase b |
| chr3_-_15264698 | 0.30 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr16_+_45739193 | 0.29 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr21_+_13861589 | 0.28 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr7_+_41887429 | 0.27 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr24_+_38306010 | 0.27 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr6_+_40661703 | 0.26 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr23_-_29668286 | 0.24 |
ENSDART00000129248
|
clstn1
|
calsyntenin 1 |
| chr4_+_11479705 | 0.23 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr6_-_32093830 | 0.23 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr10_+_21804772 | 0.23 |
ENSDART00000162194
|
pcdh1g31
|
protocadherin 1 gamma 31 |
| chr20_-_39273987 | 0.22 |
ENSDART00000127173
|
clu
|
clusterin |
| chr2_-_34555945 | 0.22 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr4_-_17785120 | 0.22 |
ENSDART00000024775
|
mybpc1
|
myosin binding protein C, slow type |
| chr15_+_25683069 | 0.21 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr5_+_27137473 | 0.21 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr6_+_12865137 | 0.21 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr16_-_25535430 | 0.20 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr14_+_19258702 | 0.20 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr22_-_15562933 | 0.20 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr17_+_33496067 | 0.20 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr9_+_6079528 | 0.20 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr21_-_26918901 | 0.20 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr2_-_14387335 | 0.19 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr10_-_7974155 | 0.19 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr15_+_11427620 | 0.19 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr18_-_898870 | 0.19 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr11_-_43200994 | 0.19 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr18_+_17583479 | 0.18 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr2_+_20332044 | 0.18 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr6_+_11249706 | 0.18 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr9_-_18568927 | 0.18 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr20_-_39273505 | 0.18 |
ENSDART00000153114
|
clu
|
clusterin |
| chr5_+_57320113 | 0.18 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr3_-_28428198 | 0.17 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr16_-_35329803 | 0.17 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr18_+_31410652 | 0.17 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr21_-_25601648 | 0.17 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr2_-_34483597 | 0.16 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr21_-_36972127 | 0.16 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr11_+_30981777 | 0.16 |
ENSDART00000148949
|
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr3_+_16724614 | 0.16 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr9_-_21918963 | 0.15 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr5_-_22882210 | 0.15 |
ENSDART00000182232
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr3_-_32169754 | 0.15 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr4_-_23963838 | 0.15 |
ENSDART00000133433
ENSDART00000132615 ENSDART00000135942 ENSDART00000139439 |
celf2
|
cugbp, Elav-like family member 2 |
| chr23_+_23182037 | 0.15 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr1_+_10019653 | 0.14 |
ENSDART00000190227
|
trim2b
|
tripartite motif containing 2b |
| chr7_+_26224211 | 0.14 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr24_+_39137001 | 0.14 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr2_+_20331445 | 0.14 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr6_+_11250033 | 0.14 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr9_+_49712868 | 0.14 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr2_-_32384683 | 0.14 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr20_+_18551657 | 0.14 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr8_+_7737062 | 0.13 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_-_48826707 | 0.13 |
ENSDART00000134711
|
svilb
|
supervillin b |
| chr18_-_2707218 | 0.13 |
ENSDART00000175226
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr20_+_16170848 | 0.13 |
ENSDART00000182115
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr8_+_31008287 | 0.13 |
ENSDART00000129882
ENSDART00000180694 |
ptgesl
|
prostaglandin E synthase 2-like |
| chr4_+_14727018 | 0.12 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr15_-_45246911 | 0.12 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr23_-_21797517 | 0.12 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr9_+_25775816 | 0.12 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr12_+_42574148 | 0.12 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr9_+_25776971 | 0.11 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr5_-_46505691 | 0.11 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr10_+_21656654 | 0.11 |
ENSDART00000160464
|
pcdh1g2
|
protocadherin 1 gamma 2 |
| chr7_-_31441420 | 0.11 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr20_+_23173710 | 0.11 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr1_+_14073891 | 0.11 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr2_-_32551178 | 0.11 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr7_+_72469724 | 0.11 |
ENSDART00000187629
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr17_-_2386569 | 0.11 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr14_-_33105434 | 0.11 |
ENSDART00000163795
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr10_+_42733210 | 0.10 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr14_-_25577094 | 0.10 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr1_-_21538673 | 0.10 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr18_+_7286788 | 0.10 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr8_+_6967108 | 0.10 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr1_+_52929185 | 0.10 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_-_40086978 | 0.10 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr12_+_9551667 | 0.10 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr20_-_22198982 | 0.10 |
ENSDART00000063578
|
srd5a3
|
steroid 5 alpha-reductase 3 |
| chr4_+_14727212 | 0.10 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr9_+_42607138 | 0.09 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr23_+_34321237 | 0.09 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr18_+_26899316 | 0.09 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr17_-_8173380 | 0.09 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr3_+_15773991 | 0.09 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr5_-_8614217 | 0.09 |
ENSDART00000164844
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr22_-_7129631 | 0.09 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr16_-_35427060 | 0.09 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr19_+_46222428 | 0.09 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr1_-_55873178 | 0.08 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr6_-_957830 | 0.08 |
ENSDART00000090019
ENSDART00000184286 |
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr11_+_36355348 | 0.08 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr14_-_23801389 | 0.08 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_+_11250316 | 0.08 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr24_+_39027481 | 0.08 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr12_+_18782821 | 0.08 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr9_-_16877456 | 0.07 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr4_+_15605844 | 0.07 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr9_+_42606681 | 0.07 |
ENSDART00000191186
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr5_+_51909740 | 0.07 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr12_+_4869248 | 0.07 |
ENSDART00000170961
|
scn4aa
|
sodium channel, voltage-gated, type IV, alpha, a |
| chr8_-_23612462 | 0.07 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr3_-_17716322 | 0.07 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr18_+_9635178 | 0.07 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr7_-_32629458 | 0.07 |
ENSDART00000001376
|
arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr8_+_10823069 | 0.07 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr25_+_28893615 | 0.07 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr7_-_22941472 | 0.07 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr12_-_24812403 | 0.07 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr7_-_51727760 | 0.07 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr7_-_30280934 | 0.07 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr21_+_27302752 | 0.07 |
ENSDART00000012855
|
sart1
|
SART1, U4/U6.U5 tri-snRNP-associated protein 1 |
| chr5_+_36654817 | 0.07 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr2_-_3403020 | 0.07 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr21_+_21201346 | 0.07 |
ENSDART00000142961
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr13_-_33667922 | 0.06 |
ENSDART00000141631
|
si:dkey-76k16.5
|
si:dkey-76k16.5 |
| chr25_-_348784 | 0.06 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr18_+_30028637 | 0.06 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr15_-_25613114 | 0.06 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr18_-_158541 | 0.06 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_-_7673376 | 0.06 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr7_+_26326462 | 0.06 |
ENSDART00000173515
|
zanl
|
zonadhesin, like |
| chr21_-_10488379 | 0.06 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr19_+_19511394 | 0.06 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr16_+_49088698 | 0.06 |
ENSDART00000182037
|
plcl2
|
phospholipase C like 2 |
| chr11_-_4175773 | 0.06 |
ENSDART00000042221
|
ccdc3b
|
coiled-coil domain containing 3b |
| chr20_-_25436389 | 0.06 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr3_-_34180364 | 0.06 |
ENSDART00000151819
ENSDART00000003133 |
yipf2
|
Yip1 domain family, member 2 |
| chr17_-_33412868 | 0.06 |
ENSDART00000187521
|
BX323819.1
|
|
| chr13_+_23162447 | 0.06 |
ENSDART00000180209
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_-_40191358 | 0.06 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr19_-_6303099 | 0.06 |
ENSDART00000133371
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr22_+_38164486 | 0.06 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr7_+_27253063 | 0.06 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr8_-_2289264 | 0.06 |
ENSDART00000189397
|
plat
|
plasminogen activator, tissue |
| chr3_-_45281350 | 0.05 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr15_+_36966369 | 0.05 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr2_+_50062165 | 0.05 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr9_-_7640692 | 0.05 |
ENSDART00000135616
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr24_-_21819010 | 0.05 |
ENSDART00000091096
|
CR352265.1
|
|
| chr22_-_29586608 | 0.05 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr19_-_43004356 | 0.05 |
ENSDART00000098101
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr20_-_47953524 | 0.05 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr3_+_25166805 | 0.05 |
ENSDART00000077493
|
TST
|
zgc:162544 |
| chr7_-_18416741 | 0.05 |
ENSDART00000097882
|
sstr1b
|
somatostatin receptor 1b |
| chr11_+_24620742 | 0.05 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr13_+_10970614 | 0.05 |
ENSDART00000058147
|
dync2li1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr13_+_29771463 | 0.05 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr9_+_13230214 | 0.05 |
ENSDART00000017932
ENSDART00000179850 |
carf
|
calcium responsive transcription factor |
| chr24_+_19578935 | 0.05 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr3_-_32603191 | 0.05 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr20_-_51727860 | 0.05 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr5_+_51026563 | 0.04 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr4_+_9536860 | 0.04 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr25_-_28768489 | 0.04 |
ENSDART00000088315
|
washc4
|
WASH complex subunit 4 |
| chr6_-_57938043 | 0.04 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr2_+_30513887 | 0.04 |
ENSDART00000137048
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr18_-_21105968 | 0.04 |
ENSDART00000100791
|
si:dkey-12e7.1
|
si:dkey-12e7.1 |
| chr10_-_42131408 | 0.04 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr9_-_4606463 | 0.04 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr24_-_32025637 | 0.04 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr5_+_33339762 | 0.04 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr20_+_28434196 | 0.04 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr16_-_30869732 | 0.04 |
ENSDART00000042716
ENSDART00000190168 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr19_+_4139065 | 0.04 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr21_-_17016788 | 0.04 |
ENSDART00000186703
|
AL844518.1
|
|
| chr16_-_36979592 | 0.04 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr1_-_23268569 | 0.04 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr11_-_14789918 | 0.04 |
ENSDART00000179670
|
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr18_-_46957840 | 0.04 |
ENSDART00000182754
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr23_-_43393589 | 0.04 |
ENSDART00000183592
|
zgc:174862
|
zgc:174862 |
| chr11_+_44236183 | 0.04 |
ENSDART00000193470
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr10_+_20364009 | 0.03 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr1_+_30100257 | 0.03 |
ENSDART00000134311
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_+_55095314 | 0.03 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr9_-_16853462 | 0.03 |
ENSDART00000160273
|
CT573248.2
|
|
| chr8_+_26254903 | 0.03 |
ENSDART00000113763
|
slc26a6
|
solute carrier family 26, member 6 |
| chr20_-_10120442 | 0.03 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr15_-_24960730 | 0.03 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr7_-_64770456 | 0.03 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr18_-_26811959 | 0.03 |
ENSDART00000086250
|
znf592
|
zinc finger protein 592 |
| chr6_+_2195625 | 0.03 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr4_-_42171503 | 0.03 |
ENSDART00000163798
|
znf1148
|
zinc finger protein 1148 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx8a+lhx8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.3 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.4 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.0 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.2 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |