Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
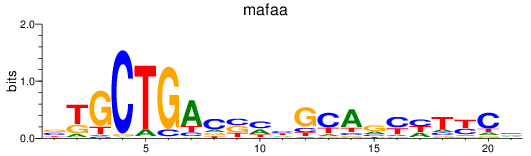
Results for mafaa
Z-value: 0.74
Transcription factors associated with mafaa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafaa
|
ENSDARG00000044155 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafaa | dr11_v1_chr6_-_32703317_32703317 | -0.73 | 1.6e-01 | Click! |
Activity profile of mafaa motif
Sorted Z-values of mafaa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39313027 | 1.24 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr13_+_4671698 | 1.08 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr4_-_8030583 | 1.08 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr10_+_19596214 | 0.89 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr2_-_22688651 | 0.82 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr19_-_5332784 | 0.72 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr7_+_27603211 | 0.69 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr8_-_50482781 | 0.64 |
ENSDART00000056361
|
ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr24_+_6353394 | 0.58 |
ENSDART00000165118
|
CR352329.1
|
|
| chr1_+_59090743 | 0.50 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr1_+_59090972 | 0.49 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr20_-_23440955 | 0.46 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr3_-_1388936 | 0.42 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr1_+_59090583 | 0.41 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr19_-_48010490 | 0.39 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr4_-_58448785 | 0.38 |
ENSDART00000172359
|
si:ch211-212k5.3
|
si:ch211-212k5.3 |
| chr6_-_33913184 | 0.37 |
ENSDART00000146373
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr4_-_8040436 | 0.36 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr3_-_10677890 | 0.35 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr24_-_6029314 | 0.34 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr21_-_1642739 | 0.31 |
ENSDART00000151118
|
zgc:152948
|
zgc:152948 |
| chr5_-_33230794 | 0.30 |
ENSDART00000144273
|
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr16_-_46567344 | 0.30 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr4_-_72468168 | 0.29 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr7_+_6969909 | 0.28 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr13_-_28272299 | 0.28 |
ENSDART00000006393
|
tlx1
|
T cell leukemia homeobox 1 |
| chr25_+_31264155 | 0.28 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr24_+_37709191 | 0.27 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr1_+_52392511 | 0.27 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr3_-_34055745 | 0.27 |
ENSDART00000151652
|
ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr8_+_52479026 | 0.26 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr11_+_25276748 | 0.26 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr19_-_3777217 | 0.25 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr5_-_23795688 | 0.25 |
ENSDART00000099084
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr25_+_3217419 | 0.23 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr25_-_27842654 | 0.23 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr6_+_42475730 | 0.23 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr13_-_50463938 | 0.22 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr21_+_18885258 | 0.22 |
ENSDART00000169078
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr4_+_56422336 | 0.22 |
ENSDART00000159185
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr8_+_43078044 | 0.22 |
ENSDART00000147872
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr23_+_3538463 | 0.20 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr16_+_4658250 | 0.19 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr3_+_26322596 | 0.19 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr16_+_42465518 | 0.19 |
ENSDART00000058699
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr3_-_62417677 | 0.18 |
ENSDART00000101888
|
sstr2b
|
somatostatin receptor 2b |
| chr23_+_12916062 | 0.18 |
ENSDART00000144268
|
si:dkey-150i13.2
|
si:dkey-150i13.2 |
| chr14_+_40641350 | 0.18 |
ENSDART00000074506
|
tnmd
|
tenomodulin |
| chr25_+_459901 | 0.17 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr18_+_33591704 | 0.17 |
ENSDART00000185631
ENSDART00000057845 |
si:dkey-47k20.5
|
si:dkey-47k20.5 |
| chr22_+_28328156 | 0.17 |
ENSDART00000166714
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr13_-_301309 | 0.16 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr21_+_45841731 | 0.16 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_-_37465517 | 0.16 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr21_-_17037907 | 0.15 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr23_+_42338325 | 0.15 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr12_-_11649690 | 0.15 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr22_+_10606863 | 0.15 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr3_+_8826905 | 0.15 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr8_-_3336273 | 0.14 |
ENSDART00000081160
|
zgc:173737
|
zgc:173737 |
| chr4_-_50926767 | 0.14 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr11_+_6115621 | 0.13 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr8_-_2591654 | 0.13 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr9_-_1639884 | 0.13 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr13_+_6295173 | 0.13 |
ENSDART00000163855
|
angpt2a
|
angiopoietin 2a |
| chr23_+_42652877 | 0.12 |
ENSDART00000035654
|
cyp2ae1
|
cytochrome P450, family 2, subfamily AE, polypeptide 1 |
| chr25_-_3217115 | 0.12 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr12_+_30789611 | 0.11 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_-_42798332 | 0.11 |
ENSDART00000189885
|
prkd3
|
protein kinase D3 |
| chr7_-_7493758 | 0.11 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr6_-_25371196 | 0.11 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr2_+_38161318 | 0.10 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr1_-_26045007 | 0.10 |
ENSDART00000129259
|
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr8_+_50534948 | 0.10 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr22_+_19283978 | 0.10 |
ENSDART00000131487
ENSDART00000143589 |
si:dkey-21e2.14
|
si:dkey-21e2.14 |
| chr10_-_57270 | 0.08 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr4_-_14486822 | 0.07 |
ENSDART00000048821
|
plxnb2a
|
plexin b2a |
| chr20_-_22476255 | 0.07 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_-_37896965 | 0.06 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr8_-_21071476 | 0.06 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr22_+_12477996 | 0.06 |
ENSDART00000177704
|
CR847870.3
|
|
| chr15_-_17138640 | 0.06 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr22_-_27709692 | 0.05 |
ENSDART00000172458
|
CR547131.1
|
|
| chr8_-_21071711 | 0.05 |
ENSDART00000111600
ENSDART00000135204 ENSDART00000131371 ENSDART00000146532 ENSDART00000137606 |
zgc:112962
|
zgc:112962 |
| chr1_-_16676177 | 0.04 |
ENSDART00000159038
|
frg1
|
FSHD region gene 1 |
| chr20_-_23946296 | 0.04 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr20_+_54323218 | 0.02 |
ENSDART00000191096
|
CABZ01077555.1
|
|
| chr17_-_20202725 | 0.01 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafaa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.7 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.1 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |