Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for mecom
Z-value: 0.72
Transcription factors associated with mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
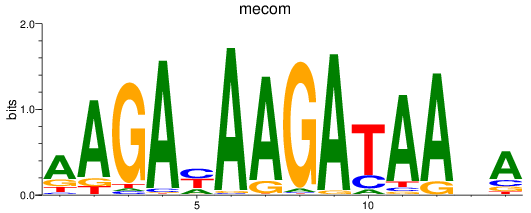
mecom
|
ENSDARG00000060808 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecom | dr11_v1_chr15_-_35410860_35410860 | 0.75 | 1.4e-01 | Click! |
Activity profile of mecom motif
Sorted Z-values of mecom motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 0.66 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr20_-_34801181 | 0.48 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr14_+_35691889 | 0.47 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr15_+_22311803 | 0.43 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr1_-_45553602 | 0.43 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr1_+_31110817 | 0.43 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr2_-_44283554 | 0.39 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr17_-_125091 | 0.38 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr16_-_52879741 | 0.34 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr10_-_31782616 | 0.34 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_-_38612230 | 0.32 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr3_-_32817274 | 0.32 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr17_+_29348013 | 0.31 |
ENSDART00000188885
ENSDART00000192005 ENSDART00000190484 ENSDART00000188565 |
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr21_+_7582036 | 0.29 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr15_-_23647078 | 0.28 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr9_+_38983895 | 0.26 |
ENSDART00000144893
|
map2
|
microtubule-associated protein 2 |
| chr2_+_38554260 | 0.26 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr19_+_16222618 | 0.26 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr11_-_3343463 | 0.25 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr13_-_11644806 | 0.25 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr21_+_5209716 | 0.25 |
ENSDART00000102539
ENSDART00000053148 ENSDART00000102536 |
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr11_+_39672874 | 0.25 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr24_-_15648636 | 0.25 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr3_+_32443395 | 0.25 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr21_-_12119711 | 0.24 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr20_+_49787584 | 0.24 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr22_-_29242347 | 0.24 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr22_+_11535131 | 0.24 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr4_+_26628822 | 0.24 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr15_-_32383529 | 0.23 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr8_-_46572298 | 0.23 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr22_+_33362552 | 0.23 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr20_+_1996202 | 0.23 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr11_-_37509001 | 0.23 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr22_-_15385442 | 0.23 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr8_+_48801402 | 0.22 |
ENSDART00000098363
|
tprg1l
|
tumor protein p63 regulated 1-like |
| chr8_-_31369161 | 0.22 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr16_+_23924040 | 0.22 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr4_-_27398385 | 0.21 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr9_+_30720048 | 0.21 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr2_-_1188707 | 0.21 |
ENSDART00000066378
|
CABZ01084564.1
|
|
| chr16_+_30959423 | 0.21 |
ENSDART00000136824
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr19_+_42693855 | 0.20 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr23_+_44745317 | 0.20 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr17_+_33495194 | 0.20 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr21_+_21326038 | 0.19 |
ENSDART00000089898
|
qpctlb
|
glutaminyl-peptide cyclotransferase-like b |
| chr24_+_41915878 | 0.19 |
ENSDART00000171523
|
TMEM200C
|
transmembrane protein 200C |
| chr2_+_2563192 | 0.19 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr18_-_21271373 | 0.19 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr5_+_44190974 | 0.19 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr17_+_13763425 | 0.19 |
ENSDART00000105175
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr5_-_10946232 | 0.19 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr12_-_6551681 | 0.18 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr15_+_16908085 | 0.18 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr11_+_35364445 | 0.18 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr13_-_33822550 | 0.18 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr10_+_22012389 | 0.18 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr17_+_26981132 | 0.18 |
ENSDART00000156672
|
si:dkey-221l4.11
|
si:dkey-221l4.11 |
| chr15_-_32383340 | 0.17 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr1_+_34181581 | 0.17 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr10_-_30785501 | 0.17 |
ENSDART00000020054
|
opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr25_-_4148719 | 0.16 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr7_+_10351038 | 0.16 |
ENSDART00000173256
|
si:cabz01029535.1
|
si:cabz01029535.1 |
| chr1_+_37752171 | 0.16 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr24_-_5889878 | 0.16 |
ENSDART00000077951
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr25_-_1235457 | 0.16 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr9_-_9348058 | 0.16 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr1_+_9071324 | 0.16 |
ENSDART00000184194
|
BX004779.2
|
|
| chr14_+_36738069 | 0.16 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr6_-_25181012 | 0.16 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr6_-_40697585 | 0.16 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr8_+_36948256 | 0.16 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr24_-_30096666 | 0.15 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr5_-_65662996 | 0.15 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr24_+_28525507 | 0.14 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr16_+_14029283 | 0.14 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr23_-_30781875 | 0.14 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr24_-_31843173 | 0.14 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr14_-_2187202 | 0.14 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr14_+_22591624 | 0.14 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr16_+_12836143 | 0.14 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr18_+_8346920 | 0.14 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr1_+_33383971 | 0.14 |
ENSDART00000150043
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr9_+_29585943 | 0.13 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr7_+_66634167 | 0.13 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr22_-_5958066 | 0.13 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr5_+_49744713 | 0.13 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr11_-_37359416 | 0.13 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr20_+_38322444 | 0.13 |
ENSDART00000161741
ENSDART00000132241 ENSDART00000148936 ENSDART00000149623 |
stum
|
stum, mechanosensory transduction mediator homolog |
| chr21_+_41743493 | 0.13 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr20_-_5369105 | 0.13 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr19_+_17259912 | 0.12 |
ENSDART00000078951
|
MANEAL
|
si:ch211-30b16.2 |
| chr22_-_17256573 | 0.12 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr6_-_6423885 | 0.12 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr18_+_16330025 | 0.12 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr14_-_47963115 | 0.11 |
ENSDART00000003826
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_38758261 | 0.11 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr13_-_39830999 | 0.11 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr3_+_37824268 | 0.11 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr3_+_32789605 | 0.11 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr3_-_18274691 | 0.11 |
ENSDART00000161140
|
BX649434.3
|
|
| chr9_+_31534601 | 0.11 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr23_+_17926279 | 0.11 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr12_+_38774860 | 0.10 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr2_-_3678029 | 0.10 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr2_+_29976419 | 0.10 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr5_-_31926906 | 0.10 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr1_+_38758445 | 0.10 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr18_+_16246806 | 0.10 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr21_-_30658509 | 0.10 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr10_+_35152928 | 0.10 |
ENSDART00000063418
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr17_-_12758171 | 0.10 |
ENSDART00000131564
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr19_+_19511394 | 0.10 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr7_+_16912837 | 0.10 |
ENSDART00000186956
|
nav2a
|
neuron navigator 2a |
| chr4_-_8902406 | 0.10 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr11_-_41130239 | 0.10 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr6_-_34008827 | 0.10 |
ENSDART00000191183
ENSDART00000003701 ENSDART00000192502 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr3_-_33437796 | 0.10 |
ENSDART00000075499
|
si:dkey-283b1.7
|
si:dkey-283b1.7 |
| chr6_-_50203682 | 0.09 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr22_-_17611742 | 0.09 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr11_-_42755108 | 0.09 |
ENSDART00000185255
|
CU459094.1
|
|
| chr10_+_41914084 | 0.09 |
ENSDART00000009227
|
orai1b
|
ORAI calcium release-activated calcium modulator 1b |
| chr13_-_39830816 | 0.09 |
ENSDART00000138981
|
zgc:171482
|
zgc:171482 |
| chr4_+_10721795 | 0.09 |
ENSDART00000136000
ENSDART00000067253 |
stab2
|
stabilin 2 |
| chr9_-_25443094 | 0.09 |
ENSDART00000105492
|
acvr2aa
|
activin A receptor type 2Aa |
| chr25_-_4235037 | 0.09 |
ENSDART00000093003
|
syt7a
|
synaptotagmin VIIa |
| chr17_-_12926456 | 0.09 |
ENSDART00000044126
|
INSM2
|
INSM transcriptional repressor 2 |
| chr23_+_42304602 | 0.09 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr2_+_51818039 | 0.09 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr9_+_13714379 | 0.09 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr22_+_14051245 | 0.09 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr10_+_34394454 | 0.09 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr20_-_5052786 | 0.09 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr25_+_2229301 | 0.09 |
ENSDART00000180198
|
pparab
|
peroxisome proliferator-activated receptor alpha b |
| chr5_-_13086616 | 0.09 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr2_-_1548330 | 0.09 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr20_+_48803248 | 0.09 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr13_+_12174937 | 0.09 |
ENSDART00000171683
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr15_+_5028608 | 0.08 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr23_-_36439961 | 0.08 |
ENSDART00000187907
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr13_-_18695289 | 0.08 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr7_+_44445595 | 0.08 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr17_-_15149192 | 0.08 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr11_-_23322182 | 0.08 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr20_-_36671660 | 0.08 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr10_-_15672862 | 0.08 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr15_-_8517555 | 0.08 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr2_+_25816843 | 0.08 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr23_+_13928346 | 0.08 |
ENSDART00000155326
|
si:dkey-90a13.10
|
si:dkey-90a13.10 |
| chr9_+_48455909 | 0.08 |
ENSDART00000167243
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr25_+_26844028 | 0.08 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr2_+_15776649 | 0.08 |
ENSDART00000156535
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr10_-_8434816 | 0.08 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr12_+_27068525 | 0.08 |
ENSDART00000188634
|
srcap
|
Snf2-related CREBBP activator protein |
| chr5_+_28161079 | 0.08 |
ENSDART00000141109
|
tacr1a
|
tachykinin receptor 1a |
| chr21_-_38031038 | 0.08 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr19_-_42557416 | 0.08 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr13_-_49707885 | 0.08 |
ENSDART00000189805
ENSDART00000128415 |
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr8_-_49499457 | 0.08 |
ENSDART00000098326
|
opn7d
|
opsin 7, group member d |
| chr11_+_4953964 | 0.08 |
ENSDART00000092552
|
ptprga
|
protein tyrosine phosphatase, receptor type, g a |
| chr19_+_37620342 | 0.07 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr20_-_7000225 | 0.07 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr13_+_30172645 | 0.07 |
ENSDART00000137114
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr25_+_25438165 | 0.07 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr5_-_5147041 | 0.07 |
ENSDART00000180236
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr2_-_10188598 | 0.07 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr23_+_30835589 | 0.07 |
ENSDART00000154145
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr25_+_33002963 | 0.07 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr24_+_23742690 | 0.07 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr9_-_9705512 | 0.07 |
ENSDART00000149021
|
gpr156
|
G protein-coupled receptor 156 |
| chr23_-_20154422 | 0.07 |
ENSDART00000132025
|
usp19
|
ubiquitin specific peptidase 19 |
| chr22_+_19111444 | 0.07 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr9_+_10014817 | 0.07 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr8_+_1187928 | 0.07 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr9_+_1138323 | 0.07 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr6_-_46861676 | 0.07 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr5_+_19165949 | 0.06 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr13_-_11971148 | 0.06 |
ENSDART00000066230
ENSDART00000185614 |
ARL3 (1 of many)
|
zgc:110197 |
| chr1_+_36771954 | 0.06 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr24_-_26460853 | 0.06 |
ENSDART00000078628
|
slc7a14b
|
solute carrier family 7, member 14b |
| chr21_+_6556635 | 0.06 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr19_+_27342479 | 0.06 |
ENSDART00000184687
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr23_+_4646194 | 0.06 |
ENSDART00000092344
|
LO017700.1
|
|
| chr21_+_15704556 | 0.06 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr9_+_10014514 | 0.06 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr13_+_29771463 | 0.06 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr9_+_31075896 | 0.06 |
ENSDART00000188042
|
clybl
|
citrate lyase beta like |
| chr4_+_27131725 | 0.06 |
ENSDART00000013083
|
brd1a
|
bromodomain containing 1a |
| chr10_+_9159279 | 0.06 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr1_-_33351780 | 0.06 |
ENSDART00000178996
|
gyg2
|
glycogenin 2 |
| chr17_-_30702411 | 0.06 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr6_-_12902772 | 0.06 |
ENSDART00000172237
|
ical1
|
islet cell autoantigen 1-like |
| chr23_+_40410644 | 0.05 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr16_+_10972733 | 0.05 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr3_+_24511959 | 0.05 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr7_+_21887307 | 0.05 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr5_+_31965845 | 0.05 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr7_-_17028015 | 0.05 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr2_-_4070850 | 0.05 |
ENSDART00000159990
|
yme1l1b
|
YME1-like 1b |
| chr22_+_35275468 | 0.05 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecom
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0021767 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.3 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |