Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
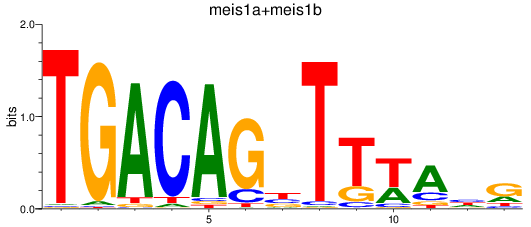
Results for meis1a+meis1b
Z-value: 0.55
Transcription factors associated with meis1a+meis1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis1a
|
ENSDARG00000002937 | Meis homeobox 1 a |
|
meis1b
|
ENSDARG00000012078 | Meis homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis1a | dr11_v1_chr1_+_51496862_51496862 | -0.79 | 1.1e-01 | Click! |
| meis1b | dr11_v1_chr13_-_5568928_5568928 | -0.71 | 1.8e-01 | Click! |
Activity profile of meis1a+meis1b motif
Sorted Z-values of meis1a+meis1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24791505 | 0.35 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr16_+_52512025 | 0.30 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr8_-_13972626 | 0.27 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr14_-_30390145 | 0.24 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr22_+_1006573 | 0.20 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr11_+_1796426 | 0.18 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr2_-_38125657 | 0.17 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr2_+_27403300 | 0.17 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr24_-_10006158 | 0.15 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr24_+_10027902 | 0.15 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr10_-_3295197 | 0.15 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr25_-_4482449 | 0.14 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr4_-_5291256 | 0.14 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr24_-_10014512 | 0.13 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr18_-_21271373 | 0.13 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr22_-_13851297 | 0.13 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr23_+_23183449 | 0.13 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr7_-_19369002 | 0.12 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr25_-_17403598 | 0.12 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr10_-_22249444 | 0.12 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr19_+_10855158 | 0.12 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr10_-_39011514 | 0.12 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr17_+_12138715 | 0.12 |
ENSDART00000112888
|
SCCPDH (1 of many)
|
zgc:174379 |
| chr9_-_37748513 | 0.12 |
ENSDART00000188967
ENSDART00000187886 |
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr23_+_23182037 | 0.12 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr19_-_3167729 | 0.11 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr8_-_17771755 | 0.11 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr17_+_47174525 | 0.11 |
ENSDART00000156831
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr18_+_1703984 | 0.11 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr21_-_16399778 | 0.11 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr1_-_20593778 | 0.11 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr24_-_9985019 | 0.11 |
ENSDART00000193536
ENSDART00000189595 |
zgc:171977
|
zgc:171977 |
| chr24_-_9979342 | 0.11 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr20_-_7176809 | 0.11 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr6_+_49926115 | 0.10 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr10_-_22831611 | 0.10 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr2_+_27855346 | 0.10 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr3_-_30609659 | 0.10 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr2_+_11028923 | 0.10 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr5_-_43682930 | 0.10 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr25_+_10830269 | 0.10 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr1_+_57331813 | 0.10 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr3_+_26734162 | 0.10 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr13_+_35765317 | 0.10 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr3_-_19133003 | 0.09 |
ENSDART00000145215
|
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr13_+_3819475 | 0.09 |
ENSDART00000139958
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr13_-_21701323 | 0.09 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr3_-_48612078 | 0.09 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr3_+_17537352 | 0.09 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr23_+_21405201 | 0.09 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr25_-_34156152 | 0.09 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr11_+_40649412 | 0.09 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr2_+_47623202 | 0.09 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr3_-_55478647 | 0.09 |
ENSDART00000153774
|
ern1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr4_-_5018705 | 0.09 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr14_+_29941445 | 0.09 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr8_+_51084993 | 0.09 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr22_+_35275468 | 0.09 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr21_+_11468934 | 0.09 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr4_-_5302162 | 0.09 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr14_-_43000836 | 0.09 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr21_+_5888641 | 0.08 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr15_+_9294620 | 0.08 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr13_+_27314795 | 0.08 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr11_-_24428237 | 0.08 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr10_-_7974155 | 0.08 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr22_+_15336752 | 0.08 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr19_-_34742440 | 0.08 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr2_-_10192459 | 0.08 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr12_-_2993095 | 0.08 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr18_+_28564640 | 0.08 |
ENSDART00000016983
|
spon1a
|
spondin 1a |
| chr17_-_43594864 | 0.08 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr24_-_17047918 | 0.08 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr7_-_65492048 | 0.08 |
ENSDART00000162381
|
dkk3a
|
dickkopf WNT signaling pathway inhibitor 3a |
| chr11_-_36957127 | 0.07 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr20_+_33519435 | 0.07 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr10_+_10387328 | 0.07 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr4_-_18281070 | 0.07 |
ENSDART00000135330
ENSDART00000179075 ENSDART00000046871 |
uhrf1bp1l
|
UHRF1 binding protein 1-like |
| chr20_-_26391958 | 0.07 |
ENSDART00000078062
|
armt1
|
acidic residue methyltransferase 1 |
| chr18_-_7810214 | 0.07 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr8_+_25173317 | 0.07 |
ENSDART00000142006
|
gpr61
|
G protein-coupled receptor 61 |
| chr16_+_26612401 | 0.07 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_-_35076730 | 0.07 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr12_+_46543572 | 0.07 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr15_-_34213898 | 0.07 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr1_-_39977551 | 0.07 |
ENSDART00000139354
|
stox2a
|
storkhead box 2a |
| chr5_-_26093945 | 0.07 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr9_+_54891962 | 0.07 |
ENSDART00000148620
ENSDART00000105620 |
si:ch211-12n8.3
|
si:ch211-12n8.3 |
| chr9_+_7358749 | 0.07 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr6_+_54154807 | 0.07 |
ENSDART00000061735
|
nudt3b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3b |
| chr8_+_14792830 | 0.07 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr17_+_28883353 | 0.07 |
ENSDART00000110322
|
prkd1
|
protein kinase D1 |
| chr9_+_38168012 | 0.07 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr7_+_27898091 | 0.07 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr2_-_32386942 | 0.07 |
ENSDART00000131408
|
ubtfl
|
upstream binding transcription factor, like |
| chr10_+_41765944 | 0.07 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr13_+_15004398 | 0.07 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr10_-_35257458 | 0.07 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr13_-_20519001 | 0.07 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr13_+_36770738 | 0.07 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr11_-_9948487 | 0.07 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr8_+_53051701 | 0.07 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr5_+_26805608 | 0.07 |
ENSDART00000111019
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr7_+_21768452 | 0.07 |
ENSDART00000127719
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr3_-_11972516 | 0.07 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr17_+_22311413 | 0.07 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr17_+_18117976 | 0.07 |
ENSDART00000123745
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr2_-_37098785 | 0.07 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr16_+_11558868 | 0.07 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr6_-_39199070 | 0.06 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr6_+_2457455 | 0.06 |
ENSDART00000157456
|
CABZ01056637.1
|
|
| chr22_-_38034852 | 0.06 |
ENSDART00000104613
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr5_-_3991655 | 0.06 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr8_+_22582146 | 0.06 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr4_-_5302866 | 0.06 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr24_+_4373033 | 0.06 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr4_-_15420452 | 0.06 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr6_-_26559921 | 0.06 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr9_-_12885201 | 0.06 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr2_-_22993601 | 0.06 |
ENSDART00000125049
ENSDART00000099735 |
mllt1b
|
MLLT1, super elongation complex subunit b |
| chr3_+_29600917 | 0.06 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr2_-_50372467 | 0.06 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr8_-_21268303 | 0.06 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr20_-_48701593 | 0.06 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr16_-_44349845 | 0.06 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr1_+_47831619 | 0.06 |
ENSDART00000110335
ENSDART00000101066 |
cfap58
|
cilia and flagella associated protein 58 |
| chr24_-_27400017 | 0.06 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr9_-_22821901 | 0.06 |
ENSDART00000101711
|
neb
|
nebulin |
| chr23_-_6660985 | 0.06 |
ENSDART00000162405
|
CR450824.2
|
|
| chr7_-_55292116 | 0.06 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr7_+_6990510 | 0.06 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr5_+_55984270 | 0.06 |
ENSDART00000047358
ENSDART00000138191 |
fkbp6
|
FK506 binding protein 6 |
| chr20_-_5291012 | 0.06 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr17_+_31185276 | 0.06 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr8_+_14959587 | 0.06 |
ENSDART00000138548
ENSDART00000041697 |
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr3_+_60716904 | 0.06 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr19_-_9503473 | 0.06 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr2_-_33676494 | 0.06 |
ENSDART00000141192
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr23_-_27589754 | 0.06 |
ENSDART00000138381
ENSDART00000133721 |
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr24_+_4373355 | 0.06 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr10_-_690072 | 0.06 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr6_+_34870374 | 0.06 |
ENSDART00000149356
|
il23r
|
interleukin 23 receptor |
| chr8_+_19621511 | 0.06 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr4_-_14191717 | 0.06 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr21_+_11749097 | 0.06 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr16_-_8132742 | 0.05 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr5_+_32009542 | 0.05 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
| chr20_-_1151265 | 0.05 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr14_+_32022272 | 0.05 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr17_-_21278582 | 0.05 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr22_-_33679277 | 0.05 |
ENSDART00000169948
|
FO904977.1
|
|
| chr22_+_35275206 | 0.05 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr2_+_15776649 | 0.05 |
ENSDART00000156535
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr25_-_22191733 | 0.05 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr10_-_24765988 | 0.05 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr25_-_22191983 | 0.05 |
ENSDART00000191181
|
pkp3a
|
plakophilin 3a |
| chr17_+_18117358 | 0.05 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr6_+_9318052 | 0.05 |
ENSDART00000162324
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_+_7557912 | 0.05 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr10_+_21576909 | 0.05 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr9_-_22822084 | 0.05 |
ENSDART00000142020
|
neb
|
nebulin |
| chr23_+_14432323 | 0.05 |
ENSDART00000178553
|
CR388373.3
|
|
| chr16_-_44649053 | 0.05 |
ENSDART00000184807
|
CR925804.2
|
|
| chr19_-_40826024 | 0.05 |
ENSDART00000131471
|
calcr
|
calcitonin receptor |
| chr3_+_29082267 | 0.05 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr2_-_55298075 | 0.05 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr2_-_41861040 | 0.05 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr8_+_19621731 | 0.05 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr7_+_61184104 | 0.05 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr3_+_12725343 | 0.05 |
ENSDART00000188583
|
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr13_-_37545487 | 0.05 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr20_-_30035326 | 0.05 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr13_+_24584401 | 0.05 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr15_-_25365319 | 0.05 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr2_-_32558795 | 0.05 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr23_+_21482571 | 0.05 |
ENSDART00000187368
|
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr20_+_25911342 | 0.05 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr11_-_16975190 | 0.04 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr8_-_41519234 | 0.04 |
ENSDART00000167283
ENSDART00000180666 |
golga1
|
golgin A1 |
| chr12_+_38695808 | 0.04 |
ENSDART00000188726
ENSDART00000003339 |
dnai2b
|
dynein, axonemal, intermediate chain 2b |
| chr25_-_28443607 | 0.04 |
ENSDART00000157243
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr19_+_7636941 | 0.04 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr11_-_42230491 | 0.04 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr25_-_16552135 | 0.04 |
ENSDART00000125836
|
si:ch211-266k8.4
|
si:ch211-266k8.4 |
| chr8_+_49570884 | 0.04 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr7_-_33960170 | 0.04 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr18_-_1228688 | 0.04 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr22_+_16418688 | 0.04 |
ENSDART00000009360
|
ankrd29
|
ankyrin repeat domain 29 |
| chr24_-_12745222 | 0.04 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr16_-_26537103 | 0.04 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr12_-_17593030 | 0.04 |
ENSDART00000168758
|
usp42
|
ubiquitin specific peptidase 42 |
| chr4_-_7875808 | 0.04 |
ENSDART00000162276
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr20_-_26491567 | 0.04 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr15_+_42933236 | 0.04 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr16_+_50089417 | 0.04 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr18_-_50579629 | 0.04 |
ENSDART00000188252
|
zgc:158464
|
zgc:158464 |
| chr24_+_9881219 | 0.04 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr7_+_6915609 | 0.04 |
ENSDART00000159213
|
ccs
|
copper chaperone for superoxide dismutase |
| chr24_-_11325849 | 0.04 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr5_+_26079178 | 0.04 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis1a+meis1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) hypothalamus cell migration(GO:0021855) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylmethionine cycle(GO:0033353) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.0 | GO:1903793 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |