Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
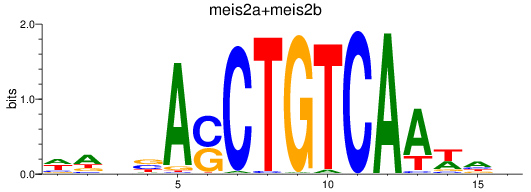
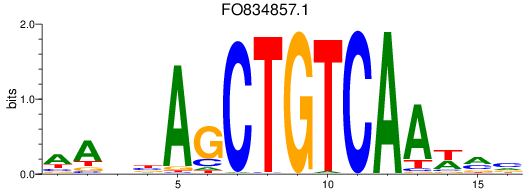
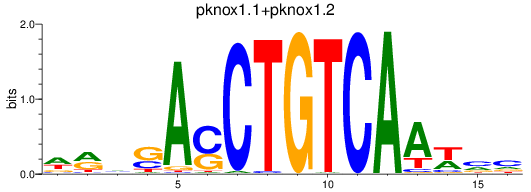
Results for meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Z-value: 0.98
Transcription factors associated with meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
|
meis2a
|
ENSDARG00000098240 | Meis homeobox 2a |
|
FO834857.1
|
ENSDARG00000112895 | homeobox protein TGIF2-like |
|
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FO834857.1 | dr11_v1_chr6_-_1820606_1820606 | -0.66 | 2.2e-01 | Click! |
| meis2b | dr11_v1_chr20_-_10118818_10118818 | -0.32 | 6.0e-01 | Click! |
| meis2a | dr11_v1_chr17_+_52822422_52822422 | -0.29 | 6.4e-01 | Click! |
| pknox1.2 | dr11_v1_chr1_-_46984142_46984142 | -0.21 | 7.3e-01 | Click! |
| pknox1.1 | dr11_v1_chr9_+_19529951_19529951 | 0.04 | 9.4e-01 | Click! |
Activity profile of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
Sorted Z-values of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_45627775 | 0.85 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr19_-_5369486 | 0.76 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr16_+_23397785 | 0.75 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr16_+_23398369 | 0.70 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr22_+_7480465 | 0.63 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr22_-_6194517 | 0.58 |
ENSDART00000134757
ENSDART00000181598 ENSDART00000129829 |
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr5_-_71722257 | 0.57 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr7_+_6879534 | 0.51 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr21_-_41025340 | 0.50 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr21_-_38153824 | 0.49 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr12_-_48168135 | 0.46 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr6_-_10809546 | 0.45 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr9_+_27876146 | 0.43 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr10_+_35417099 | 0.42 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr22_+_19188809 | 0.40 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr16_-_563235 | 0.39 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr13_-_7031033 | 0.37 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr6_+_49052741 | 0.37 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr2_-_45510699 | 0.36 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr18_+_44673990 | 0.36 |
ENSDART00000018625
|
napab
|
N-ethylmaleimide-sensitive factor attachment protein, alpha b |
| chr5_-_12587053 | 0.36 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr1_-_8653385 | 0.35 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr25_+_5068442 | 0.35 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr24_+_42148140 | 0.34 |
ENSDART00000010658
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr13_-_42749916 | 0.34 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr5_+_27432958 | 0.33 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr23_-_45705525 | 0.33 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr8_+_34731982 | 0.33 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr3_-_55099272 | 0.32 |
ENSDART00000130869
|
hbba1
|
hemoglobin, beta adult 1 |
| chr4_-_15603511 | 0.32 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr13_+_46803979 | 0.32 |
ENSDART00000159260
|
CU695232.1
|
|
| chr20_+_32152355 | 0.32 |
ENSDART00000152904
ENSDART00000139507 |
sesn1
|
sestrin 1 |
| chr16_-_563732 | 0.31 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr3_-_55104310 | 0.31 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr15_-_19724932 | 0.30 |
ENSDART00000152345
|
sytl2b
|
synaptotagmin-like 2b |
| chr3_+_24275766 | 0.30 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr18_+_29403017 | 0.30 |
ENSDART00000176966
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr12_-_36511939 | 0.30 |
ENSDART00000153759
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr24_-_36095526 | 0.30 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr7_+_55149001 | 0.30 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr9_-_31135901 | 0.30 |
ENSDART00000113027
ENSDART00000193210 ENSDART00000128896 |
si:ch211-184m13.4
|
si:ch211-184m13.4 |
| chr14_+_22022441 | 0.30 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr1_-_58000438 | 0.28 |
ENSDART00000163761
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr5_+_30741730 | 0.28 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr22_-_17677947 | 0.28 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr7_-_56766100 | 0.27 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr7_+_17816006 | 0.27 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr13_+_12671513 | 0.27 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr20_+_29634653 | 0.26 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr20_-_54564018 | 0.26 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr3_-_1400309 | 0.25 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr24_+_6353394 | 0.25 |
ENSDART00000165118
|
CR352329.1
|
|
| chr1_+_57145072 | 0.25 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr10_+_7671260 | 0.25 |
ENSDART00000157608
|
fam136a
|
family with sequence similarity 136, member A |
| chr21_-_27338639 | 0.24 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr24_+_38534550 | 0.24 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr17_-_7792376 | 0.24 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr5_-_54497319 | 0.23 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr25_-_3192405 | 0.23 |
ENSDART00000104835
|
hps5
|
Hermansky-Pudlak syndrome 5 |
| chr12_-_46176115 | 0.23 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr1_+_14253118 | 0.23 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr18_+_15758375 | 0.23 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr11_+_25459697 | 0.23 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr17_+_35362851 | 0.23 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr18_+_24562188 | 0.23 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr1_+_57187794 | 0.22 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr5_+_22307605 | 0.22 |
ENSDART00000138154
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr4_+_76692834 | 0.22 |
ENSDART00000064315
ENSDART00000137653 |
ms4a17a.1
|
membrane-spanning 4-domains, subfamily A, member 17A.1 |
| chr18_+_29402623 | 0.22 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr24_-_27430612 | 0.21 |
ENSDART00000158139
|
CR383669.2
|
|
| chr11_+_6136220 | 0.21 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr17_-_24890843 | 0.21 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr1_-_19402802 | 0.21 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr4_+_76775837 | 0.21 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr13_-_15700060 | 0.21 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr4_+_12931763 | 0.21 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr23_-_9768700 | 0.21 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr1_-_30473422 | 0.21 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr7_+_17816470 | 0.20 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_+_28160265 | 0.20 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr1_-_52437056 | 0.20 |
ENSDART00000138337
|
si:ch211-217k17.12
|
si:ch211-217k17.12 |
| chr3_+_28576173 | 0.20 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr22_-_17458070 | 0.20 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr19_+_26923274 | 0.20 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr13_+_30696286 | 0.20 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr4_+_76752707 | 0.19 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr1_-_56213723 | 0.19 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr14_-_689841 | 0.19 |
ENSDART00000125969
|
ube2ka
|
ubiquitin-conjugating enzyme E2Ka (UBC1 homolog, yeast) |
| chr23_-_13875252 | 0.19 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr14_+_29769336 | 0.19 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr3_+_29941777 | 0.19 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr23_-_20258490 | 0.19 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr9_-_48281941 | 0.19 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr3_+_22273123 | 0.18 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr24_-_37568359 | 0.18 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr11_+_44503774 | 0.18 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr9_-_7539297 | 0.18 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr19_+_7575341 | 0.18 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr19_+_7575141 | 0.18 |
ENSDART00000051528
|
s100u
|
S100 calcium binding protein U |
| chr2_+_394166 | 0.18 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr23_+_13528053 | 0.18 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr8_+_7033049 | 0.18 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr14_+_30340251 | 0.18 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr16_-_7239457 | 0.18 |
ENSDART00000148992
ENSDART00000149260 |
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr20_+_47434709 | 0.18 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr20_+_329032 | 0.18 |
ENSDART00000036635
|
fynb
|
FYN proto-oncogene, Src family tyrosine kinase b |
| chr6_+_23931236 | 0.18 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr4_-_685412 | 0.18 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr12_-_43819253 | 0.17 |
ENSDART00000160261
ENSDART00000170045 ENSDART00000159106 |
si:ch73-329n5.6
|
si:ch73-329n5.6 |
| chr25_+_5288665 | 0.17 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr10_+_44955106 | 0.17 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr7_+_51103416 | 0.17 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr21_+_43882274 | 0.17 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr25_+_19008497 | 0.17 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr16_+_33144306 | 0.17 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr21_-_32684570 | 0.17 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr16_+_20167811 | 0.17 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr22_-_21046843 | 0.16 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr20_+_1329509 | 0.16 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr12_+_18916285 | 0.16 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr19_+_7453996 | 0.16 |
ENSDART00000147491
ENSDART00000136502 |
mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr15_-_28587147 | 0.16 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr5_+_20112032 | 0.16 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr11_+_8152872 | 0.16 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr5_-_19494048 | 0.16 |
ENSDART00000098795
|
mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr11_+_26375979 | 0.16 |
ENSDART00000087652
ENSDART00000171748 ENSDART00000103513 ENSDART00000165931 ENSDART00000170043 |
cpne1
rbm12
|
copine I RNA binding motif protein 12 |
| chr23_+_6586467 | 0.15 |
ENSDART00000081763
ENSDART00000121480 |
rbm38
|
RNA binding motif protein 38 |
| chr6_-_49510553 | 0.15 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr7_+_20467549 | 0.15 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr5_-_24127310 | 0.15 |
ENSDART00000182700
ENSDART00000154313 |
capga
|
capping protein (actin filament), gelsolin-like a |
| chr16_+_5251768 | 0.15 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr1_+_58332000 | 0.15 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr10_+_11355841 | 0.15 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr24_-_40667800 | 0.15 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr10_+_26667475 | 0.15 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr22_+_25590391 | 0.15 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr24_+_10413484 | 0.15 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr20_+_27712714 | 0.15 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr16_+_17089114 | 0.15 |
ENSDART00000173223
|
cd27
|
CD27 molecule |
| chr18_+_907266 | 0.15 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr8_-_17184482 | 0.15 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr1_-_2449395 | 0.15 |
ENSDART00000103785
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr1_+_58150000 | 0.14 |
ENSDART00000137836
|
si:ch211-15j1.3
|
si:ch211-15j1.3 |
| chr19_+_48351624 | 0.14 |
ENSDART00000169729
|
sgo1
|
shugoshin 1 |
| chr19_-_38611814 | 0.14 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr12_+_34127573 | 0.14 |
ENSDART00000153413
|
cyth1b
|
cytohesin 1b |
| chr23_-_23401305 | 0.14 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr1_+_56972616 | 0.14 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr15_+_41027466 | 0.14 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr23_-_38054 | 0.14 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr3_-_16039619 | 0.14 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr19_+_7567763 | 0.14 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr21_+_45267589 | 0.14 |
ENSDART00000182963
ENSDART00000155681 ENSDART00000192632 |
tcf7
|
transcription factor 7 |
| chr8_+_25959940 | 0.13 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr19_-_12212692 | 0.13 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr8_-_4586696 | 0.13 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr6_+_38773376 | 0.13 |
ENSDART00000078128
ENSDART00000184053 |
ube3a
|
ubiquitin protein ligase E3A |
| chr1_-_44084071 | 0.13 |
ENSDART00000166912
|
vwa11
|
von Willebrand factor A domain containing 11 |
| chr24_-_27452488 | 0.13 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr13_+_24396666 | 0.13 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr14_+_35369979 | 0.13 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr24_+_23716918 | 0.13 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr14_-_25956804 | 0.13 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr22_+_980290 | 0.12 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr25_-_2723682 | 0.12 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr2_+_55365727 | 0.12 |
ENSDART00000162943
|
FP245456.1
|
|
| chr3_-_60316118 | 0.12 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr4_+_73085993 | 0.12 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr21_-_38581903 | 0.12 |
ENSDART00000148939
|
pof1b
|
POF1B, actin binding protein |
| chr16_+_33144112 | 0.12 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr7_-_38340674 | 0.12 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr13_+_45524475 | 0.12 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr21_-_34658266 | 0.12 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr6_-_42003780 | 0.12 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr1_-_12397258 | 0.12 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr13_-_37631092 | 0.12 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr5_-_37103487 | 0.11 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr21_+_17768174 | 0.11 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr1_+_53945934 | 0.11 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr23_+_24705424 | 0.11 |
ENSDART00000104029
|
c1qtnf12
|
C1q and TNF related 12 |
| chr2_+_36103309 | 0.11 |
ENSDART00000190038
|
CT867973.1
|
|
| chr12_-_3721957 | 0.11 |
ENSDART00000152433
|
vps25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr22_-_21046654 | 0.11 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr15_-_28587490 | 0.11 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr6_+_41452979 | 0.11 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr22_-_14128716 | 0.11 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr7_+_65673885 | 0.11 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr21_+_30937690 | 0.11 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr5_+_6796291 | 0.11 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr14_-_41468892 | 0.11 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr15_-_17071328 | 0.11 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr17_-_28797395 | 0.11 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr8_+_25616946 | 0.11 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr11_+_2506516 | 0.11 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr14_+_46410766 | 0.11 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr13_-_42400647 | 0.11 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr16_-_54978981 | 0.11 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr4_+_25574827 | 0.10 |
ENSDART00000187726
|
zgc:195175
|
zgc:195175 |
| chr20_-_20270191 | 0.10 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr4_-_27897160 | 0.10 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.4 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.2 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.2 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.9 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.2 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.3 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.1 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.7 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.1 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.0 | GO:0006297 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.2 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.2 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.0 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |