Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
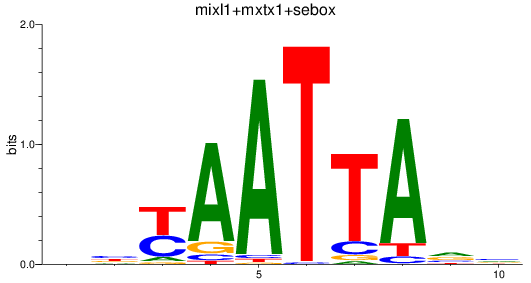
Results for mixl1+mxtx1+sebox
Z-value: 0.55
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mixl1 | dr11_v1_chr20_-_43723860_43723860 | 0.50 | 3.9e-01 | Click! |
| sebox | dr11_v1_chr5_+_67371650_67371650 | 0.15 | 8.1e-01 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_25126935 | 0.50 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr8_+_21353878 | 0.42 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr20_-_37813863 | 0.36 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_-_71389375 | 0.34 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr19_+_43297546 | 0.31 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr12_+_20352400 | 0.30 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr21_+_25777425 | 0.28 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr17_+_16046314 | 0.27 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_+_16535575 | 0.26 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr24_-_6078222 | 0.25 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr13_+_24750078 | 0.25 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr2_+_33326522 | 0.24 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr7_+_2236317 | 0.22 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr12_+_24952902 | 0.22 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr10_-_4375190 | 0.22 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr16_-_17347727 | 0.22 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr8_-_25034411 | 0.22 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr8_+_11425048 | 0.21 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr17_+_16046132 | 0.21 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_+_2631565 | 0.21 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr21_-_17296789 | 0.21 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr13_+_27232848 | 0.21 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr14_-_22113600 | 0.21 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr22_+_19552987 | 0.20 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr24_-_37640705 | 0.19 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr11_-_30634286 | 0.19 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr23_+_11285662 | 0.18 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr11_-_7261717 | 0.17 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr1_-_43727012 | 0.17 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr23_-_31913231 | 0.17 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr23_-_31913069 | 0.17 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr6_-_54826061 | 0.16 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr20_-_52902693 | 0.16 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr21_-_40173821 | 0.16 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr6_+_102506 | 0.16 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr5_+_28770273 | 0.15 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr21_+_15592426 | 0.15 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr13_+_1132261 | 0.15 |
ENSDART00000146049
|
wdr92
|
WD repeat domain 92 |
| chr15_-_43284021 | 0.14 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_+_1534644 | 0.14 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr7_+_20503344 | 0.14 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr21_-_5205617 | 0.14 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr21_-_40174647 | 0.14 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr6_+_3280939 | 0.14 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr18_+_19456648 | 0.13 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr3_-_31079186 | 0.13 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr15_-_1822548 | 0.13 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr3_+_18398876 | 0.13 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr23_+_31913292 | 0.12 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr6_-_8311044 | 0.12 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr24_-_2450597 | 0.12 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr22_-_15593824 | 0.12 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr18_+_20560442 | 0.12 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr4_+_9177997 | 0.12 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr16_+_29509133 | 0.12 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr10_+_16584382 | 0.11 |
ENSDART00000112039
|
CR790388.1
|
|
| chr19_-_19379084 | 0.11 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr14_+_6962271 | 0.11 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr4_-_9891874 | 0.11 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr12_-_35830625 | 0.11 |
ENSDART00000180028
|
CU459056.1
|
|
| chr16_+_28994709 | 0.10 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr22_-_10121880 | 0.10 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr3_+_26244353 | 0.10 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr17_-_31695217 | 0.10 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr13_-_31017960 | 0.10 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr25_+_7504314 | 0.10 |
ENSDART00000163231
|
ifitm5
|
interferon induced transmembrane protein 5 |
| chr10_-_21362071 | 0.10 |
ENSDART00000125167
|
avd
|
avidin |
| chr17_-_23616626 | 0.10 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr8_-_25033681 | 0.09 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr3_-_16719244 | 0.09 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr10_-_33297864 | 0.09 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr1_-_45616470 | 0.09 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_26244256 | 0.09 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr13_+_22295905 | 0.09 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr17_-_20118145 | 0.09 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr21_-_11654422 | 0.09 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr23_+_31912882 | 0.09 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr3_-_50443607 | 0.09 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr13_-_36535128 | 0.09 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr11_+_26604224 | 0.09 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr12_+_10631266 | 0.08 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr18_-_19456269 | 0.08 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr16_+_13883872 | 0.08 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr3_+_45365098 | 0.08 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr17_+_8799451 | 0.08 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr15_-_23376541 | 0.08 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr4_-_5455506 | 0.08 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr8_-_14126646 | 0.08 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr16_+_42471455 | 0.08 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr6_-_14292307 | 0.08 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr6_+_40922572 | 0.07 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr9_-_17417628 | 0.07 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr7_+_34592526 | 0.07 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr16_-_29387215 | 0.07 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr11_+_31864921 | 0.07 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr10_-_21362320 | 0.07 |
ENSDART00000189789
|
avd
|
avidin |
| chr18_-_40708537 | 0.07 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr16_+_21330634 | 0.07 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr11_-_45138857 | 0.07 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr16_-_43344859 | 0.07 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr1_+_52392511 | 0.07 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr11_+_36665359 | 0.07 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr1_-_18811517 | 0.07 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr17_-_25649079 | 0.07 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_-_30488063 | 0.07 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr8_-_37249813 | 0.07 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr6_+_41191482 | 0.07 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr2_-_32384683 | 0.07 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr9_-_21918963 | 0.07 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr22_-_19552796 | 0.07 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr3_-_29899172 | 0.06 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr3_+_45364849 | 0.06 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr10_-_25217347 | 0.06 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr2_-_55298075 | 0.06 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr4_+_31405646 | 0.06 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr17_-_16422654 | 0.06 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr1_-_26444075 | 0.06 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr21_-_36453594 | 0.06 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr11_-_35171162 | 0.06 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr24_+_17068724 | 0.06 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr24_-_40860603 | 0.05 |
ENSDART00000188032
|
CU633479.7
|
|
| chr11_+_12052791 | 0.05 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr14_-_33481428 | 0.05 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr24_-_27400017 | 0.05 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr23_+_39695827 | 0.05 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr15_-_41332865 | 0.05 |
ENSDART00000170014
|
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr25_-_2723682 | 0.05 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr11_-_41220794 | 0.05 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr1_+_52929185 | 0.05 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_+_21731382 | 0.05 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr5_+_4436405 | 0.05 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr23_-_25135046 | 0.04 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr8_-_16406440 | 0.04 |
ENSDART00000034004
|
faf1
|
Fas (TNFRSF6) associated factor 1 |
| chr17_-_31639845 | 0.04 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr19_+_30884706 | 0.04 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr4_-_49582108 | 0.04 |
ENSDART00000154999
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr7_+_28612671 | 0.04 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr25_+_3549584 | 0.04 |
ENSDART00000165913
|
ccdc77
|
coiled-coil domain containing 77 |
| chr11_-_1550709 | 0.04 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr19_-_5103141 | 0.03 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_50953776 | 0.03 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr19_+_15440841 | 0.03 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr17_+_43032529 | 0.03 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr5_-_14326959 | 0.03 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr24_-_30862168 | 0.03 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr3_+_28860283 | 0.03 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr22_-_2937503 | 0.03 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr23_-_19230627 | 0.03 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr3_+_32553714 | 0.03 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr23_-_17003533 | 0.03 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr8_-_37249991 | 0.03 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr14_+_23717165 | 0.03 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr15_+_5360407 | 0.03 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr25_+_11456696 | 0.03 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr22_-_20166660 | 0.03 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr21_-_39177564 | 0.03 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr8_+_37527575 | 0.02 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr5_-_37871526 | 0.02 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr3_-_39696066 | 0.02 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr16_-_16761164 | 0.02 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr24_+_19415124 | 0.02 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr20_+_52554352 | 0.02 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr6_+_50381665 | 0.02 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr7_-_51773166 | 0.02 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr4_-_71912457 | 0.02 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr15_-_18115540 | 0.02 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr7_+_23292133 | 0.01 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr13_+_4225173 | 0.01 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr3_-_39695856 | 0.01 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr21_-_22827548 | 0.01 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr7_+_19495379 | 0.01 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr3_-_30685401 | 0.01 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr10_-_32494499 | 0.01 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr22_+_31207799 | 0.01 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr17_-_41798856 | 0.01 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr4_-_43388943 | 0.01 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr1_-_55248496 | 0.01 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr19_+_10339538 | 0.01 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr8_+_23355484 | 0.01 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr15_+_31344472 | 0.01 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr19_+_15441022 | 0.01 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_+_52167986 | 0.01 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr15_+_857148 | 0.01 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr19_+_40069524 | 0.01 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr12_+_48803098 | 0.01 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr3_+_32365811 | 0.01 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr24_-_4450238 | 0.00 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr18_-_43884044 | 0.00 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr17_+_21295132 | 0.00 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr21_+_34088377 | 0.00 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr1_+_21937201 | 0.00 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr13_-_24311628 | 0.00 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr5_+_6954162 | 0.00 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr25_-_36020344 | 0.00 |
ENSDART00000181448
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr1_+_56873359 | 0.00 |
ENSDART00000152713
|
si:ch211-152f2.2
|
si:ch211-152f2.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |