Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
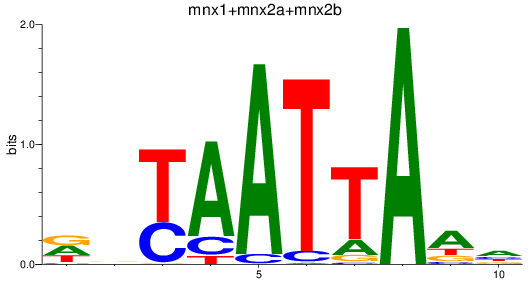
Results for mnx1+mnx2a+mnx2b
Z-value: 0.77
Transcription factors associated with mnx1+mnx2a+mnx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mnx2b
|
ENSDARG00000030350 | motor neuron and pancreas homeobox 2b |
|
mnx1
|
ENSDARG00000035984 | motor neuron and pancreas homeobox 1 |
|
mnx2a
|
ENSDARG00000042106 | motor neuron and pancreas homeobox 2a |
|
mnx1
|
ENSDARG00000109419 | motor neuron and pancreas homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mnx2b | dr11_v1_chr1_-_5455498_5455502 | -0.96 | 9.9e-03 | Click! |
| mnx2a | dr11_v1_chr9_+_7724152_7724152 | 0.78 | 1.2e-01 | Click! |
| mnx1 | dr11_v1_chr7_+_40638210_40638210 | -0.70 | 1.9e-01 | Click! |
Activity profile of mnx1+mnx2a+mnx2b motif
Sorted Z-values of mnx1+mnx2a+mnx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_36034582 | 0.84 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr10_-_8060573 | 0.53 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr3_+_13624815 | 0.51 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr1_-_43905252 | 0.50 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr10_-_21362071 | 0.47 |
ENSDART00000125167
|
avd
|
avidin |
| chr12_+_22580579 | 0.47 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_-_44801968 | 0.45 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr16_-_29387215 | 0.43 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr25_+_10410620 | 0.41 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr16_+_35916371 | 0.41 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr10_-_25217347 | 0.41 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr10_-_8046764 | 0.40 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr10_-_21362320 | 0.39 |
ENSDART00000189789
|
avd
|
avidin |
| chr17_+_43867889 | 0.35 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr22_-_24818066 | 0.35 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr18_+_15644559 | 0.35 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr17_+_16090436 | 0.34 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr14_-_22113600 | 0.34 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr11_-_25418856 | 0.34 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr20_-_37813863 | 0.33 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_-_71389375 | 0.33 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr11_+_18130300 | 0.33 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr8_+_45334255 | 0.32 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr6_-_7776612 | 0.32 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr21_-_7035599 | 0.32 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr22_+_19552987 | 0.31 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr9_-_24244383 | 0.30 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr11_+_18157260 | 0.30 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr17_+_28103675 | 0.29 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr1_+_45056371 | 0.29 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr14_+_35428152 | 0.29 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr3_-_27647845 | 0.28 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr10_-_8053385 | 0.28 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr17_+_16046314 | 0.27 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr7_+_69019851 | 0.27 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr3_-_16719244 | 0.26 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr5_-_42272517 | 0.25 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr7_+_2236317 | 0.24 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr7_-_23768234 | 0.24 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr11_-_19775182 | 0.23 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr11_-_6452444 | 0.23 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr8_-_25034411 | 0.23 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr10_+_21867307 | 0.23 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr10_+_29850330 | 0.23 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr24_-_35561672 | 0.22 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr20_-_23426339 | 0.22 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr12_-_35830625 | 0.22 |
ENSDART00000180028
|
CU459056.1
|
|
| chr11_-_25257045 | 0.22 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr6_-_43283122 | 0.22 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr8_-_24252933 | 0.21 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr1_+_44173245 | 0.21 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr13_-_31017960 | 0.21 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr15_+_6109861 | 0.21 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr22_-_36836085 | 0.21 |
ENSDART00000131451
|
cart1
|
cocaine- and amphetamine-regulated transcript 1 |
| chr18_+_19456648 | 0.20 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr23_-_40766518 | 0.20 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr8_+_2487250 | 0.20 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr2_+_37227011 | 0.20 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr13_-_27354003 | 0.19 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr24_-_6078222 | 0.19 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr5_+_66433287 | 0.19 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr21_+_1647990 | 0.19 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr20_+_27712714 | 0.19 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr4_-_9891874 | 0.19 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr22_+_10440991 | 0.19 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr18_+_3037998 | 0.18 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr6_-_39649504 | 0.18 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr20_-_40755614 | 0.18 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr18_-_19456269 | 0.18 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr8_+_2487883 | 0.18 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr20_-_48485354 | 0.17 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr23_+_11285662 | 0.17 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr20_-_45812144 | 0.17 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr11_+_31864921 | 0.16 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr6_+_3280939 | 0.16 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr11_+_12052791 | 0.16 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr19_+_43297546 | 0.16 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr10_+_16584382 | 0.16 |
ENSDART00000112039
|
CR790388.1
|
|
| chr12_+_45238292 | 0.16 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr7_-_31321027 | 0.16 |
ENSDART00000186878
|
CR356242.1
|
|
| chr18_+_20560442 | 0.16 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr4_-_16334362 | 0.15 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr20_-_43663494 | 0.15 |
ENSDART00000144564
|
BX470188.1
|
|
| chr19_+_2631565 | 0.14 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr5_+_12528693 | 0.14 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr15_+_31344472 | 0.14 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr4_+_306036 | 0.13 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr16_+_42471455 | 0.13 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr16_+_33902006 | 0.13 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr8_-_23612462 | 0.13 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr12_+_32199651 | 0.13 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr6_+_45918981 | 0.13 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr23_-_36418059 | 0.13 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr13_-_25819825 | 0.13 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr20_+_25879826 | 0.13 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr22_+_9027884 | 0.12 |
ENSDART00000171839
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr9_+_50001746 | 0.12 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr23_+_30967686 | 0.12 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr16_-_42056137 | 0.12 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr13_+_13945218 | 0.11 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr5_-_25733745 | 0.11 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr1_-_40911332 | 0.11 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr13_+_255067 | 0.11 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr9_-_50001606 | 0.11 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr17_-_37395460 | 0.11 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr3_+_27798094 | 0.11 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr8_+_41037541 | 0.11 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr11_-_25539323 | 0.11 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr12_-_6880694 | 0.10 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr15_-_4528326 | 0.10 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr13_-_42560662 | 0.10 |
ENSDART00000124898
|
CR792417.1
|
|
| chr6_+_41191482 | 0.10 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr16_-_16761164 | 0.10 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr8_-_14126646 | 0.10 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr9_+_22364997 | 0.10 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr11_-_22303678 | 0.10 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr19_+_15441022 | 0.10 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_15440841 | 0.09 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_+_52392511 | 0.09 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr8_-_30204650 | 0.09 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr22_-_10440688 | 0.09 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr1_-_45616470 | 0.09 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr17_-_32863250 | 0.09 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr15_+_5360407 | 0.09 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr2_-_38363017 | 0.09 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr2_-_51303937 | 0.09 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr17_+_7513673 | 0.09 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr5_+_25733774 | 0.09 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr25_+_13620555 | 0.09 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr19_+_30884706 | 0.09 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr13_-_8692860 | 0.09 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr3_+_32553714 | 0.08 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr7_+_39418869 | 0.08 |
ENSDART00000169195
|
CT030188.1
|
|
| chr1_-_44812014 | 0.08 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr15_+_21262917 | 0.08 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr22_+_16535575 | 0.08 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr4_-_15103646 | 0.08 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr1_-_10647484 | 0.08 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr1_+_50613868 | 0.08 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr7_-_26306546 | 0.08 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr19_-_19379084 | 0.07 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr11_-_37691449 | 0.07 |
ENSDART00000185340
|
zgc:158258
|
zgc:158258 |
| chr17_+_21295132 | 0.07 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr10_+_7593185 | 0.07 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr19_+_10339538 | 0.07 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr20_+_46492175 | 0.07 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr3_+_46635527 | 0.07 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr13_-_25719628 | 0.07 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr8_+_25034544 | 0.07 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr3_-_36210344 | 0.06 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr10_+_6884627 | 0.06 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr7_+_22585447 | 0.06 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr5_+_32847238 | 0.06 |
ENSDART00000144364
|
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr20_+_28861435 | 0.06 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr23_-_10831995 | 0.06 |
ENSDART00000142533
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr10_+_36178713 | 0.06 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr4_+_31405646 | 0.05 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr19_+_42432625 | 0.05 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr21_+_45502621 | 0.05 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr10_-_33297864 | 0.05 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr22_+_30543437 | 0.05 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr11_-_6188413 | 0.05 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr21_+_45502773 | 0.05 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr16_-_42965192 | 0.05 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr8_-_12867128 | 0.05 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_-_28671139 | 0.05 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr8_-_8446668 | 0.05 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr1_+_35985813 | 0.05 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr7_+_17229980 | 0.04 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr15_+_857148 | 0.04 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr15_-_26931541 | 0.04 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr1_-_31171242 | 0.04 |
ENSDART00000190294
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr8_+_39998467 | 0.04 |
ENSDART00000073782
ENSDART00000134452 |
ggt5a
|
gamma-glutamyltransferase 5a |
| chr23_+_28809002 | 0.04 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr7_-_44957503 | 0.04 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr5_+_37032038 | 0.04 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr14_-_32884138 | 0.04 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr3_+_32832538 | 0.04 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr12_-_13549538 | 0.04 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr18_+_8320165 | 0.04 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr2_-_22927958 | 0.04 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr20_-_34028967 | 0.04 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr13_+_21768447 | 0.04 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr5_-_13206878 | 0.03 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr20_+_28861629 | 0.03 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr12_+_48803098 | 0.03 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr17_+_14965570 | 0.03 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr11_-_39118882 | 0.03 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr19_+_42470396 | 0.03 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr7_+_48667081 | 0.03 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr20_-_52631998 | 0.03 |
ENSDART00000145283
ENSDART00000139072 |
si:ch211-221n20.1
|
si:ch211-221n20.1 |
| chr11_+_25257022 | 0.03 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_-_44065581 | 0.03 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr14_+_22113331 | 0.03 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr25_-_15268443 | 0.03 |
ENSDART00000151827
|
ccdc73
|
coiled-coil domain containing 73 |
| chr12_-_11560794 | 0.03 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr24_-_40860603 | 0.02 |
ENSDART00000188032
|
CU633479.7
|
|
| chr17_-_40956035 | 0.02 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr10_-_13343831 | 0.02 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr3_+_45368973 | 0.02 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
Network of associatons between targets according to the STRING database.
First level regulatory network of mnx1+mnx2a+mnx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0098543 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.0 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.4 | GO:0032355 | response to estradiol(GO:0032355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |