Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
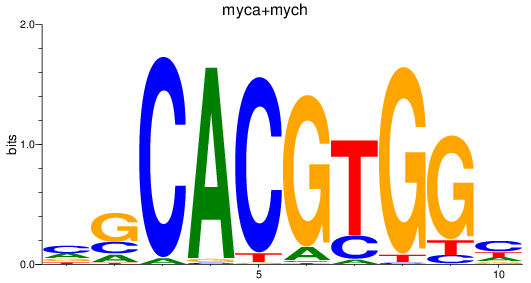
Results for myca+mych
Z-value: 0.81
Transcription factors associated with myca+mych
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myca
|
ENSDARG00000045695 | MYC proto-oncogene, bHLH transcription factor a |
|
mych
|
ENSDARG00000077473 | myelocytomatosis oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myca | dr11_v1_chr24_+_10414028_10414028 | -0.84 | 7.4e-02 | Click! |
| mych | dr11_v1_chr6_+_50451337_50451337 | 0.61 | 2.7e-01 | Click! |
Activity profile of myca+mych motif
Sorted Z-values of myca+mych motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_31555696 | 0.70 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr18_+_5490668 | 0.64 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr19_+_42983613 | 0.59 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr6_-_39764995 | 0.58 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr14_+_22076596 | 0.51 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr6_-_21189295 | 0.48 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr18_-_11729 | 0.48 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr22_+_6293563 | 0.45 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr2_-_42375275 | 0.44 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr19_-_24555935 | 0.43 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr14_+_23518110 | 0.42 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr8_+_44783424 | 0.41 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr21_-_20765338 | 0.41 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr7_+_54605640 | 0.40 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr9_+_56422517 | 0.40 |
ENSDART00000168620
|
gpr39
|
G protein-coupled receptor 39 |
| chr18_+_8917766 | 0.39 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr7_-_73752955 | 0.39 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr12_+_30586599 | 0.38 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr22_-_5323482 | 0.37 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr19_-_34117056 | 0.37 |
ENSDART00000158677
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr19_-_24555623 | 0.37 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr9_+_56422311 | 0.37 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr15_+_6661343 | 0.36 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr5_-_3927692 | 0.35 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr5_-_69314495 | 0.34 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr22_+_5120033 | 0.34 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr19_+_43119698 | 0.32 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr5_+_3927989 | 0.32 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr12_-_33359052 | 0.32 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_11969603 | 0.32 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr12_-_33359654 | 0.32 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr13_-_12021566 | 0.32 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr11_-_37995501 | 0.31 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr16_-_38629208 | 0.31 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr22_-_607812 | 0.31 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr20_+_46660339 | 0.30 |
ENSDART00000016530
|
adcy3b
|
adenylate cyclase 3b |
| chr16_-_42186093 | 0.30 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr17_+_8212477 | 0.30 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr9_-_12888082 | 0.30 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr22_+_5118361 | 0.29 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr7_+_67467702 | 0.29 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr10_-_39130839 | 0.29 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr12_-_9132682 | 0.29 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr16_+_25245857 | 0.29 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr20_+_25445826 | 0.28 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr23_+_32499916 | 0.28 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr11_-_44194132 | 0.28 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr18_-_11595567 | 0.28 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr7_+_20031202 | 0.28 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr21_+_6114709 | 0.27 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr19_-_46957968 | 0.27 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr5_-_8765428 | 0.27 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr12_-_33357655 | 0.27 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_+_26059677 | 0.26 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr18_+_14277003 | 0.26 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr3_-_37148594 | 0.26 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr20_-_33705044 | 0.25 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr21_+_26071874 | 0.24 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr13_-_31370184 | 0.24 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr20_+_37294112 | 0.24 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr21_+_21796663 | 0.23 |
ENSDART00000003518
|
neu3.2
|
sialidase 3 (membrane sialidase), tandem duplicate 2 |
| chr20_-_52882881 | 0.23 |
ENSDART00000111078
|
wu:fi04e12
|
wu:fi04e12 |
| chr10_+_22034477 | 0.23 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr14_+_16151636 | 0.23 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr6_-_41536323 | 0.23 |
ENSDART00000113157
|
hemk1
|
HemK methyltransferase family member 1 |
| chr7_-_23745984 | 0.22 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr9_+_12887491 | 0.22 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr14_+_16151368 | 0.22 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr7_+_20344032 | 0.22 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr16_+_40954481 | 0.21 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr20_-_33704753 | 0.21 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr21_-_30082414 | 0.21 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr7_+_6652967 | 0.21 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr4_-_2945306 | 0.21 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr5_-_32338866 | 0.21 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_30779575 | 0.20 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr25_-_36248053 | 0.20 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr13_-_3516473 | 0.19 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr21_-_22122312 | 0.18 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr8_+_15277874 | 0.18 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr6_-_54078623 | 0.18 |
ENSDART00000154076
|
hyal1
|
hyaluronoglucosaminidase 1 |
| chr6_-_52348562 | 0.18 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr2_-_6182098 | 0.17 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr12_+_27117609 | 0.17 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr11_+_3959495 | 0.17 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr19_+_20201254 | 0.17 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_-_45370200 | 0.17 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr24_-_42090635 | 0.17 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr16_-_8520474 | 0.17 |
ENSDART00000137365
|
grb10b
|
growth factor receptor-bound protein 10b |
| chr23_+_38159715 | 0.17 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr23_+_43870886 | 0.17 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr20_-_53996193 | 0.17 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr20_+_54666222 | 0.17 |
ENSDART00000166592
|
CABZ01087948.1
|
|
| chr20_-_54259780 | 0.16 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr5_-_33280699 | 0.16 |
ENSDART00000183838
|
kyat1
|
kynurenine aminotransferase 1 |
| chr11_-_16975190 | 0.16 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr12_-_30359031 | 0.16 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr6_-_52796212 | 0.16 |
ENSDART00000154133
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr3_-_18805225 | 0.16 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_+_20030888 | 0.16 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr1_-_19648227 | 0.16 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr2_+_9946121 | 0.16 |
ENSDART00000100696
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr11_+_6010177 | 0.16 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr11_+_6009984 | 0.15 |
ENSDART00000185680
|
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr6_-_39080630 | 0.15 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr13_-_9061944 | 0.15 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr20_+_54079341 | 0.15 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr23_-_38160024 | 0.15 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr4_-_25271455 | 0.15 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr7_-_58843193 | 0.15 |
ENSDART00000167231
|
mrpl15
|
mitochondrial ribosomal protein L15 |
| chr3_+_1150348 | 0.15 |
ENSDART00000148524
|
nol12
|
nucleolar protein 12 |
| chr17_-_49407091 | 0.15 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr4_+_2620751 | 0.15 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr15_-_17099560 | 0.15 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr22_-_26834043 | 0.14 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr16_+_16968682 | 0.14 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr5_-_30978381 | 0.14 |
ENSDART00000127787
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr8_-_19904124 | 0.14 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr20_-_40725142 | 0.14 |
ENSDART00000181279
|
cx32.2
|
connexin 32.2 |
| chr16_-_54455573 | 0.14 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr17_+_24109012 | 0.14 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr4_-_14192254 | 0.14 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr9_+_42270043 | 0.13 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr24_+_32668675 | 0.13 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr17_-_27382826 | 0.13 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr7_-_2116512 | 0.13 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr7_+_17973269 | 0.13 |
ENSDART00000079969
|
kcnk7
|
potassium channel, subfamily K, member 7 |
| chr7_-_50410524 | 0.13 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr13_-_25408387 | 0.13 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr19_+_20201593 | 0.13 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_+_36627099 | 0.12 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr6_+_3717613 | 0.12 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr1_-_55750208 | 0.12 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr13_-_35892051 | 0.12 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr16_+_16969060 | 0.12 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr6_-_18531349 | 0.12 |
ENSDART00000160693
ENSDART00000169780 |
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr25_+_3231812 | 0.12 |
ENSDART00000163647
|
metap2b
|
methionyl aminopeptidase 2b |
| chr12_-_20350629 | 0.12 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr10_+_428269 | 0.12 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr24_+_23791758 | 0.12 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr14_+_94946 | 0.12 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr10_+_35002786 | 0.11 |
ENSDART00000099552
|
exosc8
|
exosome component 8 |
| chr18_-_5850683 | 0.11 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr1_-_40015782 | 0.11 |
ENSDART00000157425
ENSDART00000159238 |
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr20_+_25626479 | 0.11 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr19_-_35229336 | 0.11 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr20_-_26531850 | 0.11 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr10_+_29849977 | 0.11 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr1_+_29664336 | 0.11 |
ENSDART00000088290
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr7_+_1550966 | 0.11 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr1_+_45925365 | 0.11 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr21_-_35853245 | 0.11 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr16_-_10223741 | 0.11 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr1_-_54972170 | 0.11 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr3_+_28939759 | 0.10 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr10_+_29850330 | 0.10 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr14_-_5407118 | 0.10 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr14_-_12822 | 0.10 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr8_-_52859301 | 0.10 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr24_+_16905188 | 0.10 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr12_+_13091842 | 0.10 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr22_-_36690742 | 0.10 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr11_+_25485774 | 0.10 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr17_-_29771639 | 0.10 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr13_+_25761279 | 0.10 |
ENSDART00000177818
ENSDART00000002863 |
neurog3
|
neurogenin 3 |
| chr9_+_38158570 | 0.10 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr14_+_94603 | 0.10 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_55914268 | 0.10 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr13_-_4223955 | 0.10 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr2_-_48171112 | 0.10 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr2_-_17393971 | 0.09 |
ENSDART00000100201
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr15_-_43978141 | 0.09 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr1_+_44390470 | 0.09 |
ENSDART00000100321
|
timm10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr20_+_38257766 | 0.09 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr21_-_41147818 | 0.09 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr14_+_5385855 | 0.09 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr3_+_58379450 | 0.09 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr2_-_24369087 | 0.09 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr24_-_38192003 | 0.09 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr14_+_14836468 | 0.09 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr5_-_16475374 | 0.09 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_-_54195084 | 0.09 |
ENSDART00000171811
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr22_-_5099824 | 0.09 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr23_+_36122058 | 0.08 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr21_-_30408775 | 0.08 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr6_-_39653972 | 0.08 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr13_+_30572172 | 0.08 |
ENSDART00000010052
ENSDART00000144417 |
ppifa
|
peptidylprolyl isomerase Fa |
| chr21_+_38638979 | 0.08 |
ENSDART00000143373
|
rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr24_+_34069675 | 0.08 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr24_-_16904803 | 0.08 |
ENSDART00000165972
ENSDART00000149974 ENSDART00000149706 |
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr3_-_41791178 | 0.08 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr13_-_35907768 | 0.08 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr13_+_48359573 | 0.08 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr15_-_1484795 | 0.08 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr9_-_30259295 | 0.08 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr2_+_30547018 | 0.08 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr9_+_41821613 | 0.08 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr24_+_36317544 | 0.08 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr13_+_30912385 | 0.08 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr1_+_45925150 | 0.08 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
Network of associatons between targets according to the STRING database.
First level regulatory network of myca+mych
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.4 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.1 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.7 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.2 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.3 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.2 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0046385 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.2 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0061333 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.7 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.2 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |