Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for myog
Z-value: 1.20
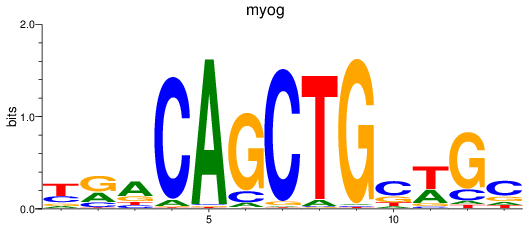
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr11_v1_chr11_-_22599584_22599584 | 0.74 | 1.6e-01 | Click! |
Activity profile of myog motif
Sorted Z-values of myog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 1.16 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr20_-_27325258 | 1.16 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr21_+_27382893 | 1.05 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr11_-_101758 | 0.88 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr7_+_29951997 | 0.80 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr8_-_51404806 | 0.72 |
ENSDART00000060625
|
lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr23_-_45405968 | 0.66 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr3_+_57991074 | 0.57 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_-_58130703 | 0.55 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr20_+_35382482 | 0.55 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr6_-_32703317 | 0.53 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr2_+_42191592 | 0.53 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr12_-_4756478 | 0.51 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr7_+_39444843 | 0.50 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr14_-_32258759 | 0.48 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr17_-_4318393 | 0.48 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr22_+_20720808 | 0.47 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr11_+_7158723 | 0.46 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr3_+_26145013 | 0.45 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr12_-_11570 | 0.45 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr6_-_21492752 | 0.44 |
ENSDART00000006843
ENSDART00000171479 |
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr20_-_32112818 | 0.43 |
ENSDART00000142653
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr25_-_19433244 | 0.42 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr21_-_30254185 | 0.42 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr19_-_10425140 | 0.42 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr13_-_31622195 | 0.41 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr18_+_5549672 | 0.41 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr17_+_27434626 | 0.40 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr16_-_14074594 | 0.40 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr3_-_36115339 | 0.39 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr10_-_34741738 | 0.38 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr12_+_24562667 | 0.38 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr24_-_33703504 | 0.37 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr15_-_26636826 | 0.37 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr25_+_35683956 | 0.36 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr11_-_3343463 | 0.35 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr13_+_912123 | 0.35 |
ENSDART00000169931
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr23_-_31633201 | 0.34 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr19_-_5254699 | 0.34 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr10_+_466926 | 0.34 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr4_-_77624155 | 0.34 |
ENSDART00000099761
|
si:ch211-250m6.7
|
si:ch211-250m6.7 |
| chr21_+_5129513 | 0.34 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr16_-_13789908 | 0.33 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr3_+_31953145 | 0.33 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr10_+_1668106 | 0.33 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr16_-_42894628 | 0.32 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr3_-_62380146 | 0.32 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr23_-_3674443 | 0.31 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr17_-_34963575 | 0.31 |
ENSDART00000145664
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr18_+_15271993 | 0.31 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr8_+_44926946 | 0.31 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr18_+_5454341 | 0.30 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr7_+_46003449 | 0.30 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr3_-_36260102 | 0.30 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr11_-_45171139 | 0.30 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr20_+_19066596 | 0.30 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr2_-_21335131 | 0.29 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr16_+_41873708 | 0.29 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr16_+_26766423 | 0.29 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr25_+_17689565 | 0.29 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr9_-_18568927 | 0.29 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr3_+_26144765 | 0.29 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr20_-_36887176 | 0.28 |
ENSDART00000143515
ENSDART00000146994 |
txlnba
|
taxilin beta a |
| chr2_-_42393590 | 0.28 |
ENSDART00000135529
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr20_-_36887516 | 0.28 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr3_+_35298078 | 0.28 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr21_-_20329541 | 0.28 |
ENSDART00000166049
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr8_-_1051438 | 0.27 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr1_+_49266886 | 0.27 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr1_+_41666611 | 0.27 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr6_-_32093830 | 0.26 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr5_+_42467867 | 0.26 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr4_-_5597802 | 0.26 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr6_-_38418862 | 0.26 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr10_-_24362775 | 0.25 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr18_+_39028417 | 0.25 |
ENSDART00000148428
|
myo5aa
|
myosin VAa |
| chr25_-_32869794 | 0.25 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr23_+_22656477 | 0.25 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr9_-_327901 | 0.25 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr9_-_100579 | 0.25 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr5_-_46273938 | 0.25 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr18_+_23218980 | 0.25 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr21_+_40417152 | 0.25 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr20_-_54381034 | 0.25 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr6_-_42003780 | 0.24 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr10_-_34871737 | 0.24 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr23_-_32162810 | 0.23 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr11_-_55224 | 0.23 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr13_-_36911118 | 0.23 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr5_-_48307804 | 0.23 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr20_+_19066858 | 0.23 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr3_-_51912019 | 0.23 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr18_+_49411417 | 0.23 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr2_+_42177113 | 0.23 |
ENSDART00000056441
|
tmeff1a
|
transmembrane protein with EGF-like and two follistatin-like domains 1a |
| chr20_+_1278184 | 0.23 |
ENSDART00000039616
|
lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr13_+_3954715 | 0.23 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr6_+_44812803 | 0.23 |
ENSDART00000169713
|
chl1b
|
cell adhesion molecule L1-like b |
| chr2_+_5621529 | 0.22 |
ENSDART00000144187
|
fgf12a
|
fibroblast growth factor 12a |
| chr7_-_28147838 | 0.22 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr5_-_13167097 | 0.22 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr16_+_29630965 | 0.21 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr22_-_3595439 | 0.21 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr9_-_48281941 | 0.21 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr4_-_2350371 | 0.21 |
ENSDART00000166274
|
phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr1_-_471925 | 0.21 |
ENSDART00000152684
|
zgc:92518
|
zgc:92518 |
| chr6_+_13730522 | 0.20 |
ENSDART00000153524
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr14_-_32405387 | 0.20 |
ENSDART00000184647
|
fgf13a
|
fibroblast growth factor 13a |
| chr9_-_30145080 | 0.20 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr23_-_18057553 | 0.20 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr4_+_10066840 | 0.20 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr14_-_29906209 | 0.20 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr13_+_3954540 | 0.20 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr25_+_36674715 | 0.20 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr16_+_39159752 | 0.20 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr6_+_21095918 | 0.20 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr14_+_7939398 | 0.20 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr24_+_20575259 | 0.19 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr12_+_24561832 | 0.19 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr19_-_30904590 | 0.19 |
ENSDART00000137633
|
si:ch211-194e15.5
|
si:ch211-194e15.5 |
| chr3_+_23488652 | 0.19 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr20_-_40319890 | 0.19 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr5_-_38342992 | 0.19 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr24_-_5910811 | 0.19 |
ENSDART00000163012
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr13_+_40019001 | 0.19 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr18_+_27732285 | 0.19 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr16_-_31188715 | 0.19 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr8_-_4596662 | 0.19 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr22_-_17574511 | 0.19 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr15_-_12500938 | 0.19 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr5_+_31779911 | 0.19 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr14_+_7939216 | 0.19 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr23_-_15284757 | 0.18 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr5_-_38384755 | 0.18 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr11_+_36477481 | 0.18 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr17_-_22001303 | 0.18 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr18_+_23249519 | 0.18 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr1_+_39553040 | 0.18 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr17_+_52822831 | 0.18 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr5_+_36768674 | 0.17 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr7_-_69636502 | 0.17 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr17_-_722218 | 0.17 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr20_-_18736281 | 0.17 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr1_+_41690402 | 0.17 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr5_+_20147830 | 0.17 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr11_-_36963988 | 0.17 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr12_-_25380028 | 0.17 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr24_-_6501211 | 0.17 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr3_-_46410387 | 0.17 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr5_+_36781732 | 0.17 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr20_+_46560258 | 0.17 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr8_-_26609259 | 0.16 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr3_+_46724528 | 0.16 |
ENSDART00000181358
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr19_+_42770041 | 0.16 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr15_+_30917282 | 0.16 |
ENSDART00000129474
|
omgb
|
oligodendrocyte myelin glycoprotein b |
| chr24_-_9300160 | 0.16 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_+_11120532 | 0.16 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr8_+_1651821 | 0.16 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr23_-_3409140 | 0.16 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr10_-_42776344 | 0.16 |
ENSDART00000190653
ENSDART00000130229 |
wdr45
|
WD repeat domain 45 |
| chr14_-_27121854 | 0.16 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr21_+_45366229 | 0.16 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr12_+_47446158 | 0.16 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr16_+_54592907 | 0.16 |
ENSDART00000172113
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr8_-_22639794 | 0.16 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr1_+_37195919 | 0.16 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr18_-_1228688 | 0.15 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr6_-_16393491 | 0.15 |
ENSDART00000179312
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr14_-_18671334 | 0.15 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr18_-_85294 | 0.15 |
ENSDART00000044387
|
gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr17_-_39786222 | 0.15 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr10_-_690072 | 0.15 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr10_+_15777258 | 0.15 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr16_+_36768674 | 0.15 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr1_-_20271138 | 0.15 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr19_-_8604429 | 0.15 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr23_-_44219902 | 0.15 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr15_+_25489406 | 0.15 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr13_-_40499296 | 0.15 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr20_+_54036981 | 0.15 |
ENSDART00000060455
|
wdr20b
|
WD repeat domain 20b |
| chr16_-_29458806 | 0.14 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr22_+_18929412 | 0.14 |
ENSDART00000161598
ENSDART00000166650 ENSDART00000015951 ENSDART00000105392 ENSDART00000131131 |
bsg
|
basigin |
| chr5_+_42912966 | 0.14 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr23_-_30781875 | 0.14 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr9_+_34334156 | 0.14 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr6_-_18250857 | 0.14 |
ENSDART00000159486
|
anapc11
|
APC11 anaphase promoting complex subunit 11 homolog (yeast) |
| chr2_-_31735142 | 0.14 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr5_-_38384289 | 0.14 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr4_+_3980247 | 0.14 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr20_+_23625387 | 0.14 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr1_+_37196106 | 0.14 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr21_-_38717854 | 0.14 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr16_-_41465542 | 0.14 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr12_+_40945319 | 0.14 |
ENSDART00000183435
|
CDH18
|
cadherin 18 |
| chr16_-_9980402 | 0.14 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr9_-_43071519 | 0.13 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr18_+_25003207 | 0.13 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr17_-_45254585 | 0.13 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr14_-_46832825 | 0.13 |
ENSDART00000149571
|
ldb2a
|
LIM domain binding 2a |
| chr12_+_17504559 | 0.13 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr8_+_36948256 | 0.13 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.4 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.3 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.3 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.5 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 0.1 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.3 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.0 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |