Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
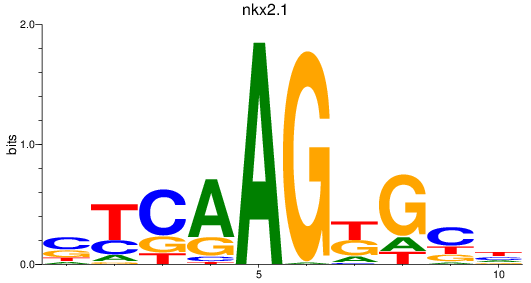
Results for nkx2.1
Z-value: 0.79
Transcription factors associated with nkx2.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.1
|
ENSDARG00000019835 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.1 | dr11_v1_chr17_+_38262408_38262408 | -0.43 | 4.7e-01 | Click! |
Activity profile of nkx2.1 motif
Sorted Z-values of nkx2.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_28674739 | 0.96 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr5_+_51597677 | 0.81 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr5_+_51594209 | 0.73 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr17_+_20605013 | 0.59 |
ENSDART00000156878
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr4_-_67799941 | 0.50 |
ENSDART00000185830
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr25_+_25124684 | 0.50 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr3_+_24207243 | 0.49 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr2_+_26237322 | 0.46 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr7_+_48555400 | 0.46 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr7_+_48555626 | 0.43 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr6_-_39051319 | 0.35 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr22_+_39096911 | 0.35 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chrM_+_10590 | 0.33 |
ENSDART00000093615
|
mt-nd3
|
NADH dehydrogenase 3, mitochondrial |
| chr23_-_33558161 | 0.33 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr2_+_26240339 | 0.27 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr7_+_25053331 | 0.25 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr18_+_808911 | 0.25 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr12_+_34953038 | 0.23 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr1_+_55662491 | 0.21 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr22_-_24248420 | 0.21 |
ENSDART00000165433
|
rgs2
|
regulator of G protein signaling 2 |
| chr7_+_34602991 | 0.20 |
ENSDART00000073447
|
fhod1
|
formin homology 2 domain containing 1 |
| chr3_-_31019715 | 0.18 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr8_-_36469117 | 0.18 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr21_-_2348838 | 0.17 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr22_+_38762693 | 0.16 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr13_-_36579086 | 0.15 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr12_-_30583668 | 0.15 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr2_-_8609653 | 0.14 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr11_-_24063196 | 0.13 |
ENSDART00000036513
|
trib3
|
tribbles pseudokinase 3 |
| chr4_-_20314749 | 0.13 |
ENSDART00000066894
ENSDART00000188123 |
dcp1b
|
decapping mRNA 1B |
| chr3_+_17933132 | 0.12 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr4_-_14531687 | 0.12 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr11_-_36040549 | 0.11 |
ENSDART00000112684
|
setmar
|
SET domain and mariner transposase fusion gene |
| chr7_-_30639385 | 0.11 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr5_-_35456269 | 0.11 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr6_+_38880166 | 0.11 |
ENSDART00000019939
ENSDART00000144286 |
bin2b
|
bridging integrator 2b |
| chr11_+_43403525 | 0.10 |
ENSDART00000180683
|
vipb
|
vasoactive intestinal peptide b |
| chr15_-_434503 | 0.10 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr3_-_8510201 | 0.09 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr10_-_16062565 | 0.08 |
ENSDART00000188728
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr2_-_59345920 | 0.08 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr1_+_46026457 | 0.08 |
ENSDART00000132705
|
si:ch211-138g9.2
|
si:ch211-138g9.2 |
| chr22_-_5724085 | 0.08 |
ENSDART00000110526
ENSDART00000140905 |
pgbd4
|
piggyBac transposable element derived 4 |
| chr13_+_27040887 | 0.08 |
ENSDART00000132714
|
soul2
|
heme-binding protein soul2 |
| chr1_+_27977297 | 0.07 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr25_+_19485198 | 0.06 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr1_-_59571758 | 0.06 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr6_-_46875310 | 0.06 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr9_+_7724152 | 0.06 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr6_+_38879961 | 0.06 |
ENSDART00000184798
|
bin2b
|
bridging integrator 2b |
| chr7_+_48675347 | 0.06 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr4_+_76748500 | 0.05 |
ENSDART00000075607
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr18_-_39321484 | 0.05 |
ENSDART00000077694
|
leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr11_-_8271374 | 0.05 |
ENSDART00000168253
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr21_+_34088377 | 0.05 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr7_-_48733662 | 0.05 |
ENSDART00000191675
|
traf6
|
TNF receptor-associated factor 6 |
| chr15_-_23482088 | 0.04 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr1_-_43862638 | 0.04 |
ENSDART00000145044
|
tacr3a
|
tachykinin receptor 3a |
| chr1_-_58000438 | 0.04 |
ENSDART00000163761
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr16_+_5539189 | 0.04 |
ENSDART00000178106
|
CR848788.2
|
|
| chr17_+_23975762 | 0.03 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr11_+_12879635 | 0.03 |
ENSDART00000182515
ENSDART00000081296 |
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr4_-_50926767 | 0.03 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr11_+_583725 | 0.03 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr1_-_23294753 | 0.03 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr3_+_22335030 | 0.03 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr8_-_44298964 | 0.03 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr15_-_4053149 | 0.03 |
ENSDART00000189076
|
CR626884.4
|
|
| chr12_+_20506197 | 0.02 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr4_-_2052687 | 0.02 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr1_+_55583116 | 0.02 |
ENSDART00000152163
|
adgre19
|
adhesion G protein-coupled receptor E19 |
| chr9_-_7089303 | 0.02 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr14_-_33478963 | 0.02 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr7_-_59123066 | 0.02 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr7_+_42461850 | 0.02 |
ENSDART00000190350
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr21_-_2162850 | 0.02 |
ENSDART00000159731
|
gb:ai877918
|
expressed sequence AI877918 |
| chr20_+_46255057 | 0.02 |
ENSDART00000100536
|
taar14i
|
trace amine associated receptor 14i |
| chr12_+_30788912 | 0.02 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_+_18531932 | 0.01 |
ENSDART00000165271
|
suz12b
|
SUZ12 polycomb repressive complex 2b subunit |
| chr7_-_3895071 | 0.01 |
ENSDART00000175200
|
si:dkey-88n24.11
|
si:dkey-88n24.11 |
| chr20_+_9128829 | 0.01 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr7_+_25825667 | 0.01 |
ENSDART00000149835
|
mtm1
|
myotubularin 1 |
| chr11_-_1956204 | 0.01 |
ENSDART00000185541
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr18_-_22701800 | 0.00 |
ENSDART00000135098
|
si:ch73-113g13.3
|
si:ch73-113g13.3 |
| chr2_-_52020049 | 0.00 |
ENSDART00000128198
ENSDART00000135331 |
scn12aa
|
sodium channel, voltage gated, type XII, alpha a |
| chr3_-_7464250 | 0.00 |
ENSDART00000159873
|
znf1001
|
zinc finger protein 1001 |
| chr14_-_16476863 | 0.00 |
ENSDART00000089021
|
canx
|
calnexin |
| chr17_+_24597001 | 0.00 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0048241 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |