Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
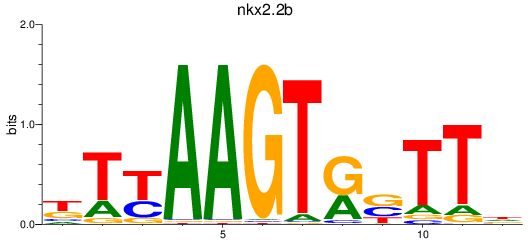
Results for nkx2.2b
Z-value: 0.94
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr11_v1_chr20_+_48782068_48782068 | -0.81 | 9.7e-02 | Click! |
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_33063083 | 0.98 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr2_-_24269911 | 0.78 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr5_+_43782267 | 0.75 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr2_-_24270062 | 0.75 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr22_+_11775269 | 0.67 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr8_+_25302172 | 0.65 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr7_-_16562200 | 0.54 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_-_30636163 | 0.53 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr13_+_8840772 | 0.52 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr16_-_51288178 | 0.50 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr1_-_43905252 | 0.50 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr2_-_28396993 | 0.49 |
ENSDART00000188170
|
CABZ01056052.1
|
|
| chr20_+_53577502 | 0.48 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr17_+_20616196 | 0.47 |
ENSDART00000153515
|
si:ch73-306e8.2
|
si:ch73-306e8.2 |
| chr22_-_17631675 | 0.46 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr3_-_33970190 | 0.45 |
ENSDART00000151238
|
ighj2-5
|
ighj2-5 |
| chr14_+_17376940 | 0.45 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr11_-_25257595 | 0.43 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr17_+_20616576 | 0.42 |
ENSDART00000153561
|
si:ch73-306e8.2
|
si:ch73-306e8.2 |
| chr24_+_38197852 | 0.41 |
ENSDART00000142949
|
igl3v4
|
immunoglobulin light 3 variable 4 |
| chr17_+_6538733 | 0.41 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr2_-_10877228 | 0.40 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr22_+_835728 | 0.40 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr7_+_21275152 | 0.39 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr24_+_38205798 | 0.39 |
ENSDART00000141532
|
igl3v1
|
immunoglobulin light 3 variable 1 |
| chr19_-_48330287 | 0.39 |
ENSDART00000162244
|
si:ch73-359m17.7
|
si:ch73-359m17.7 |
| chr3_-_27647845 | 0.38 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr2_+_26498446 | 0.38 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr24_-_30263301 | 0.38 |
ENSDART00000162328
|
snx7
|
sorting nexin 7 |
| chr6_+_13201358 | 0.37 |
ENSDART00000190290
|
CT009620.1
|
|
| chr9_+_41156287 | 0.37 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr23_-_39784368 | 0.36 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr22_+_19453822 | 0.35 |
ENSDART00000141371
|
si:dkey-78l4.6
|
si:dkey-78l4.6 |
| chr7_+_50849142 | 0.35 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr12_+_6002715 | 0.35 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr5_+_1983495 | 0.35 |
ENSDART00000179860
|
CT573103.2
|
|
| chr23_+_38245610 | 0.34 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr4_+_2230701 | 0.34 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr11_+_14284866 | 0.33 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr25_+_10416583 | 0.33 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr24_-_36680261 | 0.32 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr4_-_22311610 | 0.32 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr8_+_1170015 | 0.32 |
ENSDART00000081457
ENSDART00000164116 |
ccl27a
|
chemokine (C-C motif) ligand 27a |
| chr7_-_8309505 | 0.32 |
ENSDART00000182530
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr21_+_19547806 | 0.32 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr14_-_10617923 | 0.31 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr16_+_24842662 | 0.31 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr11_+_14286160 | 0.30 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr2_+_48282590 | 0.29 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr6_+_49255706 | 0.29 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr8_+_52442622 | 0.29 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr8_-_45760087 | 0.29 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr2_-_58183499 | 0.28 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr11_+_3308656 | 0.28 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr13_-_22699024 | 0.28 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr5_+_1877464 | 0.27 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr7_+_12950507 | 0.27 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr3_+_12829566 | 0.27 |
ENSDART00000157672
|
si:ch211-8c17.2
|
si:ch211-8c17.2 |
| chr19_+_9344171 | 0.27 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr13_-_30996072 | 0.26 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr2_+_36121373 | 0.26 |
ENSDART00000187002
|
CT867973.2
|
|
| chr16_-_11798994 | 0.26 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr1_-_49558223 | 0.26 |
ENSDART00000138510
|
si:ch211-281g13.4
|
si:ch211-281g13.4 |
| chr8_-_38317914 | 0.25 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr4_-_9891874 | 0.25 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr11_-_7411436 | 0.25 |
ENSDART00000167312
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr1_+_1915967 | 0.25 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr7_-_22790630 | 0.24 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr21_+_76739 | 0.24 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr3_+_19216567 | 0.24 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr5_-_37117778 | 0.24 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr2_+_36114194 | 0.24 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr3_-_11008532 | 0.24 |
ENSDART00000165086
|
CR382337.3
|
|
| chr22_-_6941098 | 0.24 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr12_-_30558694 | 0.24 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr18_-_43866001 | 0.23 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr19_+_40856807 | 0.23 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr21_-_36619599 | 0.23 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr11_+_31285127 | 0.23 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr9_-_18911608 | 0.23 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr23_-_24394719 | 0.23 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr21_+_33454147 | 0.22 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr5_-_38777852 | 0.22 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr16_+_35887868 | 0.22 |
ENSDART00000169677
ENSDART00000170772 |
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr4_+_6032640 | 0.21 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr1_+_55703120 | 0.21 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr9_+_18023288 | 0.21 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr5_+_3927989 | 0.21 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr5_-_30380593 | 0.21 |
ENSDART00000148039
|
snx19a
|
sorting nexin 19a |
| chr1_+_17900306 | 0.21 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr15_+_37589698 | 0.21 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr24_-_26484298 | 0.21 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr2_+_1487118 | 0.21 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr19_+_4066449 | 0.21 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr13_-_40401870 | 0.21 |
ENSDART00000128951
|
nkx3.3
|
NK3 homeobox 3 |
| chr2_-_51772438 | 0.21 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_+_883678 | 0.21 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr15_+_31526225 | 0.21 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr1_+_55478653 | 0.21 |
ENSDART00000152441
|
adgre9
|
adhesion G protein-coupled receptor E9 |
| chr8_-_38105053 | 0.21 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr20_-_39333657 | 0.20 |
ENSDART00000153720
ENSDART00000142164 |
ccl38.1
|
chemokine (C-C motif) ligand 38, duplicate 1 |
| chr9_+_1313418 | 0.20 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr12_-_30548244 | 0.20 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr14_+_1170968 | 0.20 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr20_-_32045057 | 0.20 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr4_-_25215968 | 0.20 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr23_+_19813677 | 0.20 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr7_-_17627320 | 0.19 |
ENSDART00000101701
|
nitr6a
|
novel immune-type receptor 6a |
| chr2_+_1486822 | 0.19 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr20_+_492529 | 0.19 |
ENSDART00000141709
|
FO904980.1
|
|
| chr13_+_18515126 | 0.19 |
ENSDART00000044697
|
tlr4ba
|
toll-like receptor 4b, duplicate a |
| chr15_-_1198886 | 0.19 |
ENSDART00000063285
|
lxn
|
latexin |
| chr9_-_9998087 | 0.19 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr8_-_49304602 | 0.19 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr3_-_34107685 | 0.19 |
ENSDART00000151130
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr22_-_4892937 | 0.19 |
ENSDART00000147002
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr15_-_20468302 | 0.19 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr7_+_71586485 | 0.18 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr3_+_28581397 | 0.18 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr23_+_35847538 | 0.18 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr9_+_33222911 | 0.18 |
ENSDART00000135387
|
si:ch211-125e6.14
|
si:ch211-125e6.14 |
| chr4_-_12477224 | 0.18 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr5_+_28497956 | 0.18 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr20_-_5267600 | 0.18 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr1_+_55723870 | 0.18 |
ENSDART00000146614
|
adgre17
|
adhesion G protein-coupled receptor E17 |
| chr1_-_55248496 | 0.18 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr9_+_40546677 | 0.18 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr5_+_42141917 | 0.18 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr7_+_29992889 | 0.18 |
ENSDART00000055936
|
isl2b
|
ISL LIM homeobox 2b |
| chr13_-_865193 | 0.17 |
ENSDART00000187053
|
AL929536.7
|
|
| chr8_-_43847138 | 0.17 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr14_+_12169979 | 0.17 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr12_+_23812530 | 0.17 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr5_-_9073433 | 0.17 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr7_+_65564163 | 0.17 |
ENSDART00000082679
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr1_+_5485799 | 0.17 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr12_-_28794957 | 0.17 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr21_-_40835069 | 0.16 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr4_-_16644708 | 0.16 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr3_-_16110100 | 0.16 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr6_-_7439490 | 0.16 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr8_-_50482781 | 0.16 |
ENSDART00000056361
|
ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr7_+_35268054 | 0.16 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr25_+_4635355 | 0.16 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr6_-_39024538 | 0.16 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr23_-_6765653 | 0.16 |
ENSDART00000192310
|
FP102169.1
|
|
| chr14_-_733565 | 0.15 |
ENSDART00000158097
|
tlr1
|
toll-like receptor 1 |
| chr4_+_69823638 | 0.15 |
ENSDART00000165786
|
znf1087
|
zinc finger protein 1087 |
| chr17_+_19481049 | 0.15 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr17_+_7513673 | 0.15 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr3_+_4403980 | 0.15 |
ENSDART00000156881
|
CR774195.1
|
|
| chr2_+_11028923 | 0.15 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr6_-_26895314 | 0.15 |
ENSDART00000134259
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr8_+_20624510 | 0.15 |
ENSDART00000138604
|
nfic
|
nuclear factor I/C |
| chr20_-_34090740 | 0.15 |
ENSDART00000062539
ENSDART00000008140 |
pdcb
|
phosducin b |
| chr12_+_36109507 | 0.15 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_37638873 | 0.15 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr7_-_71585065 | 0.15 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr14_-_9400552 | 0.14 |
ENSDART00000129485
ENSDART00000131341 |
tbx22
|
T-box 22 |
| chr20_+_53368611 | 0.14 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr25_+_19008497 | 0.14 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr3_+_60761811 | 0.14 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr12_-_3705862 | 0.14 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr16_+_5926520 | 0.14 |
ENSDART00000162229
|
ulk4
|
unc-51 like kinase 4 |
| chr2_-_15324837 | 0.14 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr14_-_25956804 | 0.14 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr12_-_4301234 | 0.14 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr10_-_22117121 | 0.14 |
ENSDART00000149470
|
si:ch73-111e15.1
|
si:ch73-111e15.1 |
| chr4_+_58576146 | 0.14 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr10_+_8481660 | 0.14 |
ENSDART00000109559
|
BX682234.1
|
|
| chr6_-_1762191 | 0.14 |
ENSDART00000167928
|
orc4
|
origin recognition complex, subunit 4 |
| chr18_-_6766354 | 0.14 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr13_-_25819825 | 0.14 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr23_-_4409668 | 0.13 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr2_-_24402341 | 0.13 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr22_-_7700179 | 0.13 |
ENSDART00000145047
|
si:ch211-59h6.1
|
si:ch211-59h6.1 |
| chr12_+_30726425 | 0.13 |
ENSDART00000153275
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr21_+_5272059 | 0.13 |
ENSDART00000141981
|
loxhd1a
|
lipoxygenase homology domains 1a |
| chr4_+_31405646 | 0.13 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr14_+_34952883 | 0.13 |
ENSDART00000182812
|
il12ba
|
interleukin 12Ba |
| chr11_+_2398843 | 0.13 |
ENSDART00000126761
|
pip4k2cb
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma b |
| chr14_-_46173265 | 0.13 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr15_-_37589600 | 0.13 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr15_+_37972671 | 0.13 |
ENSDART00000166764
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr16_+_20915319 | 0.13 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr13_-_43238578 | 0.13 |
ENSDART00000161017
|
eif3s6ip
|
eukaryotic translation initiation factor 3, subunit 6 interacting protein |
| chr12_+_41348969 | 0.12 |
ENSDART00000171352
|
si:ch211-27e6.1
|
si:ch211-27e6.1 |
| chr4_-_16876281 | 0.12 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr11_+_1565806 | 0.12 |
ENSDART00000137479
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr16_+_23913943 | 0.12 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr10_-_25343521 | 0.12 |
ENSDART00000166348
ENSDART00000009477 |
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr1_-_354115 | 0.12 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr7_+_32901658 | 0.12 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr10_+_44373349 | 0.12 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr1_+_23784905 | 0.12 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr20_+_25712276 | 0.12 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr13_+_44857087 | 0.12 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr20_-_50049099 | 0.12 |
ENSDART00000123634
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr7_+_57108823 | 0.12 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr21_-_14310159 | 0.12 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.8 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.5 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.4 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.2 | GO:0006706 | steroid catabolic process(GO:0006706) cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.1 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |