Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
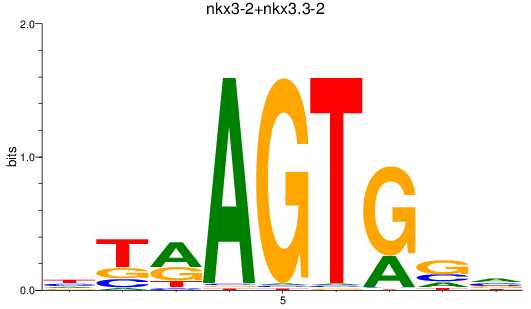
Results for nkx3-2+nkx3.3-2
Z-value: 0.52
Transcription factors associated with nkx3-2+nkx3.3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-2
|
ENSDARG00000037639 | NK3 homeobox 2 |
|
nkx3.3-2
|
ENSDARG00000069327 | NK3 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3.3 | dr11_v1_chr13_-_40401870_40401870 | 0.50 | 4.0e-01 | Click! |
| nkx3.2 | dr11_v1_chr14_-_215051_215051 | -0.37 | 5.4e-01 | Click! |
Activity profile of nkx3-2+nkx3.3-2 motif
Sorted Z-values of nkx3-2+nkx3.3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_19188809 | 0.45 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr12_+_22580579 | 0.42 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr1_-_48933 | 0.36 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr15_-_31027112 | 0.36 |
ENSDART00000100185
|
lgals9l4
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 4 |
| chr19_+_19989380 | 0.31 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr1_+_1789357 | 0.30 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr17_+_6563307 | 0.29 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr6_+_42918933 | 0.26 |
ENSDART00000064896
|
gnat1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr2_+_39618951 | 0.25 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr16_-_24815091 | 0.24 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr18_-_20458412 | 0.23 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr6_+_13201358 | 0.21 |
ENSDART00000190290
|
CT009620.1
|
|
| chr11_+_14333441 | 0.21 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr3_-_34060153 | 0.20 |
ENSDART00000151610
|
ighv1-1
|
immunoglobulin heavy variable 1-1 |
| chr16_-_16522013 | 0.20 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr16_+_49005321 | 0.19 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr3_-_32603191 | 0.19 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr4_+_72668095 | 0.18 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr16_+_26612401 | 0.18 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr5_-_26764880 | 0.18 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr20_-_30377221 | 0.17 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr10_+_13209580 | 0.17 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr6_-_40038543 | 0.17 |
ENSDART00000154792
|
si:dkey-197j19.6
|
si:dkey-197j19.6 |
| chr7_-_4843401 | 0.16 |
ENSDART00000137489
|
si:dkey-83f18.8
|
si:dkey-83f18.8 |
| chr24_+_17005647 | 0.15 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr19_+_32855139 | 0.15 |
ENSDART00000052082
|
rpl30
|
ribosomal protein L30 |
| chr19_+_43604643 | 0.14 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr21_-_11791909 | 0.14 |
ENSDART00000180893
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr17_+_51746830 | 0.14 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr5_-_42083363 | 0.13 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr7_-_23777445 | 0.13 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr18_+_16943911 | 0.13 |
ENSDART00000157609
|
si:dkey-8l13.5
|
si:dkey-8l13.5 |
| chr1_-_51710225 | 0.13 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr13_+_23093743 | 0.12 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr4_-_71436560 | 0.12 |
ENSDART00000166884
|
si:dkey-82i20.1
|
si:dkey-82i20.1 |
| chr5_+_61843752 | 0.12 |
ENSDART00000130940
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr3_-_34054081 | 0.12 |
ENSDART00000151590
|
ighv1-2
|
immunoglobulin heavy variable 1-2 |
| chr21_-_11996769 | 0.12 |
ENSDART00000143537
|
zgc:64106
|
zgc:64106 |
| chr8_-_48847772 | 0.12 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr8_+_25299069 | 0.11 |
ENSDART00000114676
|
gstm.2
|
glutathione S-transferase mu tandem duplicate 2 |
| chr15_-_41245962 | 0.11 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr7_+_54222156 | 0.11 |
ENSDART00000165201
ENSDART00000158518 |
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr2_-_51644044 | 0.11 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr15_+_15173611 | 0.11 |
ENSDART00000155267
|
si:ch211-149e23.4
|
si:ch211-149e23.4 |
| chr23_+_25292147 | 0.11 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr2_+_49631850 | 0.11 |
ENSDART00000114274
ENSDART00000114516 |
si:dkey-53k12.18
|
si:dkey-53k12.18 |
| chr25_+_19670273 | 0.10 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr8_+_24281512 | 0.10 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr16_+_27444098 | 0.09 |
ENSDART00000157690
|
invs
|
inversin |
| chr8_-_4971205 | 0.09 |
ENSDART00000184250
|
tmem230b
|
transmembrane protein 230b |
| chr18_-_22094102 | 0.09 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr4_+_17327704 | 0.09 |
ENSDART00000016075
ENSDART00000133160 |
nup37
|
nucleoporin 37 |
| chr5_-_26765188 | 0.09 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr7_+_20917966 | 0.09 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr4_-_75812937 | 0.09 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr6_-_39198912 | 0.09 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr20_-_25643667 | 0.09 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr3_-_16413606 | 0.08 |
ENSDART00000127309
ENSDART00000017172 ENSDART00000136465 |
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr18_-_26781616 | 0.08 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr6_+_374875 | 0.08 |
ENSDART00000171698
|
si:zfos-169g10.3
|
si:zfos-169g10.3 |
| chr12_+_19030391 | 0.08 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr3_-_16039619 | 0.07 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr21_+_4388489 | 0.07 |
ENSDART00000144555
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr15_+_5360407 | 0.07 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr12_+_17754859 | 0.07 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr2_+_42135719 | 0.07 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr4_-_13931508 | 0.07 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr19_-_3777217 | 0.06 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr4_+_71382288 | 0.06 |
ENSDART00000181926
|
si:ch211-76m11.8
|
si:ch211-76m11.8 |
| chr8_-_4031121 | 0.06 |
ENSDART00000169474
ENSDART00000163754 |
mtmr3
|
myotubularin related protein 3 |
| chr22_+_9431853 | 0.06 |
ENSDART00000181028
|
si:ch211-11p18.6
|
si:ch211-11p18.6 |
| chr3_-_41791178 | 0.06 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr25_+_16356083 | 0.06 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr7_-_6441865 | 0.06 |
ENSDART00000172831
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr16_-_22585289 | 0.06 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr25_+_11456696 | 0.06 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr5_-_8907819 | 0.05 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr16_+_28994709 | 0.05 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr11_-_36474306 | 0.05 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr22_-_26514386 | 0.05 |
ENSDART00000125628
|
CR759791.1
|
|
| chr10_+_24048883 | 0.05 |
ENSDART00000149265
|
gbe1a
|
glucan (1,4-alpha-), branching enzyme 1a |
| chr5_-_23117078 | 0.05 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr15_-_36347858 | 0.05 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr23_+_12840080 | 0.04 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr5_+_60928576 | 0.04 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr22_-_27113332 | 0.04 |
ENSDART00000146951
ENSDART00000178855 |
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr15_+_24676905 | 0.04 |
ENSDART00000078014
ENSDART00000143137 |
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr3_-_41795917 | 0.04 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr15_-_36727462 | 0.04 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr21_+_34814444 | 0.04 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr21_+_4403155 | 0.04 |
ENSDART00000161623
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr12_-_13729263 | 0.04 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr4_+_45357558 | 0.03 |
ENSDART00000150769
|
si:ch211-162i8.5
|
si:ch211-162i8.5 |
| chr10_-_29831944 | 0.03 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr6_+_58622831 | 0.03 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr22_-_20386982 | 0.03 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr12_-_16636627 | 0.03 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr8_-_17987547 | 0.02 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr18_-_38216584 | 0.02 |
ENSDART00000144622
|
si:dkey-10o6.2
|
si:dkey-10o6.2 |
| chr20_+_25568694 | 0.02 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr9_-_9225980 | 0.02 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr11_-_5953636 | 0.02 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr4_+_71018579 | 0.02 |
ENSDART00000186727
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr7_+_44715224 | 0.02 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr14_-_1200854 | 0.02 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr22_+_9922301 | 0.02 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr11_-_12634017 | 0.02 |
ENSDART00000158286
ENSDART00000193090 |
CR450764.1
|
|
| chr2_-_55797318 | 0.01 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr1_-_14258409 | 0.01 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr15_-_35246742 | 0.01 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr18_-_27858123 | 0.01 |
ENSDART00000142068
|
iqcg
|
IQ motif containing G |
| chr8_+_28547687 | 0.01 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr1_-_28861226 | 0.01 |
ENSDART00000075502
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr20_+_6773790 | 0.00 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-2+nkx3.3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |