Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
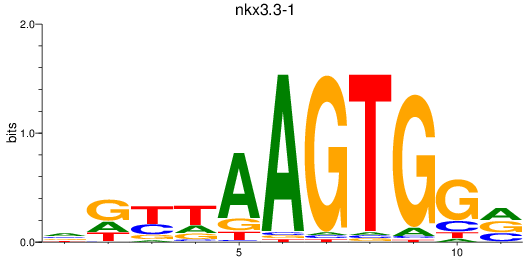
Results for nkx3.3-1
Z-value: 0.91
Transcription factors associated with nkx3.3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3.3-1
|
ENSDARG00000110589 | NK3 homeobox 3 |
Activity profile of nkx3.3-1 motif
Sorted Z-values of nkx3.3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_16022211 | 0.85 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr5_+_2815021 | 0.80 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr8_-_14554785 | 0.61 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_+_37978501 | 0.59 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr7_-_23777445 | 0.56 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr8_-_50147948 | 0.55 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr12_+_31713239 | 0.55 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr15_+_45563491 | 0.53 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr22_+_38194151 | 0.53 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr2_+_51028269 | 0.52 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr15_+_5973909 | 0.48 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr25_+_28282274 | 0.47 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr3_+_29640837 | 0.46 |
ENSDART00000132298
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr4_-_14642379 | 0.45 |
ENSDART00000114977
|
si:ch211-127b11.1
|
si:ch211-127b11.1 |
| chr21_+_26612777 | 0.41 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr3_-_32912787 | 0.39 |
ENSDART00000156149
|
g6pcb
|
glucose-6-phosphatase b, catalytic subunit |
| chr15_+_46606090 | 0.38 |
ENSDART00000020921
|
CABZ01044048.1
|
|
| chr23_+_25292147 | 0.35 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr5_+_1983495 | 0.34 |
ENSDART00000179860
|
CT573103.2
|
|
| chr22_-_26236188 | 0.34 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr4_-_14649158 | 0.31 |
ENSDART00000145737
|
si:dkey-183c2.4
|
si:dkey-183c2.4 |
| chr14_+_26719691 | 0.31 |
ENSDART00000078522
ENSDART00000172927 |
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr21_+_5589923 | 0.31 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr12_+_19030391 | 0.30 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr24_+_15670013 | 0.30 |
ENSDART00000185826
|
CU929414.1
|
|
| chr15_-_3277635 | 0.30 |
ENSDART00000189094
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr19_+_43604643 | 0.29 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr1_-_54706039 | 0.29 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr11_+_42556395 | 0.29 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr24_-_25166720 | 0.28 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr19_+_30990815 | 0.28 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr5_-_54712159 | 0.27 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr2_-_23600821 | 0.27 |
ENSDART00000146217
|
BX677666.1
|
|
| chr25_+_11456696 | 0.26 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr9_-_43073960 | 0.26 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr16_-_38609146 | 0.26 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr5_-_23855447 | 0.26 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr18_+_29156827 | 0.25 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr19_+_30990129 | 0.25 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr21_-_28523548 | 0.24 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr5_-_8907819 | 0.24 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr2_-_23538794 | 0.24 |
ENSDART00000143857
|
si:dkey-58b18.10
|
si:dkey-58b18.10 |
| chr16_+_16977786 | 0.23 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr10_+_25726694 | 0.23 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr4_+_16715267 | 0.23 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr10_-_39052264 | 0.23 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr22_+_32228882 | 0.23 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr15_-_41245962 | 0.23 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr25_-_19666107 | 0.22 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr9_-_49531762 | 0.22 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr21_-_41873065 | 0.22 |
ENSDART00000014538
|
endou2
|
endonuclease, polyU-specific 2 |
| chr4_-_72468168 | 0.22 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr21_+_21812311 | 0.22 |
ENSDART00000151253
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr9_+_8380728 | 0.21 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr5_+_13870340 | 0.21 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr25_+_2361721 | 0.21 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr19_+_791538 | 0.21 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr4_-_11098539 | 0.21 |
ENSDART00000135511
|
si:dkey-21h14.12
|
si:dkey-21h14.12 |
| chr23_-_17003533 | 0.21 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr3_+_11926030 | 0.20 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr24_-_26484298 | 0.20 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr9_-_53666031 | 0.20 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr23_+_32044410 | 0.20 |
ENSDART00000048628
|
mylk2
|
myosin light chain kinase 2 |
| chr9_+_15893093 | 0.20 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr15_+_33991928 | 0.20 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr24_-_33308045 | 0.20 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr21_-_43094210 | 0.20 |
ENSDART00000144747
|
si:ch73-68b22.2
|
si:ch73-68b22.2 |
| chr5_-_57204352 | 0.19 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr2_+_53114476 | 0.19 |
ENSDART00000080850
ENSDART00000104187 |
CR384061.1
|
|
| chr14_-_1280907 | 0.19 |
ENSDART00000186150
|
CABZ01081490.1
|
|
| chr25_-_37284370 | 0.19 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr7_+_17393794 | 0.19 |
ENSDART00000059673
|
nitr7b
|
novel immune-type receptor 7b |
| chr19_-_34970312 | 0.19 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr5_-_12063381 | 0.19 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr18_+_33100606 | 0.19 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr3_-_34066548 | 0.18 |
ENSDART00000151302
|
ighv9-2
|
immunoglobulin heavy variable 9-2 |
| chr4_-_8030583 | 0.18 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr25_+_28772632 | 0.18 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr21_+_18925318 | 0.18 |
ENSDART00000136182
|
si:ch211-222n4.2
|
si:ch211-222n4.2 |
| chr13_-_40237121 | 0.18 |
ENSDART00000145635
|
loxl4
|
lysyl oxidase-like 4 |
| chr10_+_10387328 | 0.18 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr25_+_10547228 | 0.18 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr17_+_6563307 | 0.18 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr2_+_30281444 | 0.18 |
ENSDART00000172756
|
si:dkey-82k12.13
|
si:dkey-82k12.13 |
| chr10_+_10386435 | 0.18 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr6_+_58622831 | 0.18 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr22_+_9872323 | 0.18 |
ENSDART00000129240
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr16_-_35394422 | 0.18 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr7_+_7048245 | 0.18 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr5_+_37890521 | 0.18 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_+_16452912 | 0.17 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr25_+_20091021 | 0.17 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr13_-_50463938 | 0.17 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr23_-_39959784 | 0.17 |
ENSDART00000115046
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr1_+_56779495 | 0.17 |
ENSDART00000192871
|
FO680692.1
|
|
| chr1_+_7988052 | 0.17 |
ENSDART00000167552
|
CR855320.2
|
|
| chr3_-_41795917 | 0.16 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr11_-_21031773 | 0.16 |
ENSDART00000065985
|
fmoda
|
fibromodulin a |
| chr18_+_24562188 | 0.16 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr16_+_4658250 | 0.16 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr7_-_59311165 | 0.16 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr8_+_24281512 | 0.16 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr4_+_17327704 | 0.15 |
ENSDART00000016075
ENSDART00000133160 |
nup37
|
nucleoporin 37 |
| chr12_-_44016898 | 0.15 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr22_+_1330477 | 0.15 |
ENSDART00000157567
|
CU207221.2
|
|
| chr12_+_29236274 | 0.15 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr22_+_9922301 | 0.15 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr18_-_33080454 | 0.14 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr11_-_12634017 | 0.14 |
ENSDART00000158286
ENSDART00000193090 |
CR450764.1
|
|
| chr3_+_2971852 | 0.14 |
ENSDART00000059271
|
BX004816.1
|
|
| chr11_+_31680513 | 0.14 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr16_+_3982590 | 0.14 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr3_-_34052882 | 0.14 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr7_+_12835048 | 0.14 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr2_+_30948503 | 0.13 |
ENSDART00000149923
|
myom1a
|
myomesin 1a (skelemin) |
| chr15_+_15771418 | 0.13 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr3_-_2091029 | 0.13 |
ENSDART00000141464
|
si:dkey-88j15.4
|
si:dkey-88j15.4 |
| chr15_+_37972671 | 0.13 |
ENSDART00000166764
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr21_+_5993188 | 0.13 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr19_-_6840506 | 0.13 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr6_+_3280939 | 0.13 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr1_-_58887610 | 0.13 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr16_+_27444098 | 0.13 |
ENSDART00000157690
|
invs
|
inversin |
| chr16_-_22585289 | 0.12 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr3_-_5393909 | 0.12 |
ENSDART00000031885
|
si:ch73-106l15.2
|
si:ch73-106l15.2 |
| chr4_-_30349370 | 0.12 |
ENSDART00000161790
|
znf1047
|
zinc finger protein 1047 |
| chr5_+_8492640 | 0.12 |
ENSDART00000171701
|
osmr
|
oncostatin M receptor |
| chr13_-_6038477 | 0.12 |
ENSDART00000166130
|
CABZ01033394.1
|
|
| chr4_-_49875682 | 0.12 |
ENSDART00000185921
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr16_+_38240027 | 0.12 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr2_+_22496973 | 0.12 |
ENSDART00000159638
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr5_-_23783739 | 0.12 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr7_-_17267950 | 0.12 |
ENSDART00000122171
|
nitr10a
|
novel immune-type receptor 10a |
| chr4_+_61995745 | 0.12 |
ENSDART00000171539
|
CT990567.1
|
|
| chr18_+_2593756 | 0.12 |
ENSDART00000158022
|
p2ry2.3
|
purinergic receptor P2Y2, tandem duplicate 3 |
| chr21_+_34814444 | 0.12 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr19_+_7810028 | 0.12 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr13_-_35892051 | 0.12 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr7_+_50796422 | 0.12 |
ENSDART00000123868
|
dthd1
|
death domain containing 1 |
| chr20_-_33675676 | 0.12 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr4_-_10958074 | 0.11 |
ENSDART00000150478
|
si:ch211-161n3.4
|
si:ch211-161n3.4 |
| chr8_-_51753604 | 0.11 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr2_+_45593783 | 0.11 |
ENSDART00000148037
|
fndc7rs2
|
fibronectin type III domain containing 7, related sequence 2 |
| chr22_+_19522982 | 0.11 |
ENSDART00000192428
ENSDART00000190812 |
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr9_-_32656656 | 0.11 |
ENSDART00000078399
|
gzm3
|
granzyme 3, tandem duplicate 1 |
| chr22_-_13350240 | 0.11 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr5_+_58550291 | 0.11 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr4_-_10835620 | 0.11 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr7_-_70283372 | 0.11 |
ENSDART00000112969
|
adgra3
|
adhesion G protein-coupled receptor A3 |
| chr1_-_50791280 | 0.11 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr4_+_70343496 | 0.11 |
ENSDART00000123293
ENSDART00000158476 |
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr18_-_34171280 | 0.11 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr3_-_22356568 | 0.11 |
ENSDART00000058507
|
ifnphi2
|
interferon phi 2 |
| chr5_+_44846280 | 0.11 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr4_+_72668095 | 0.11 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr5_-_24124118 | 0.11 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr12_-_16990896 | 0.10 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr1_-_35113974 | 0.10 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr12_+_17754859 | 0.10 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr13_+_33462232 | 0.10 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr15_-_40041455 | 0.10 |
ENSDART00000099359
|
lpar5b
|
lysophosphatidic acid receptor 5b |
| chr4_-_71708567 | 0.10 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr23_-_46040618 | 0.10 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr12_+_22580579 | 0.10 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr9_-_34882516 | 0.10 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr17_-_2811992 | 0.10 |
ENSDART00000067543
|
gpr65
|
G protein-coupled receptor 65 |
| chr15_+_17756113 | 0.10 |
ENSDART00000155197
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr7_-_7984015 | 0.09 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr18_+_13182528 | 0.09 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr17_+_48164536 | 0.09 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr4_+_53146060 | 0.09 |
ENSDART00000124912
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr1_+_18863060 | 0.09 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr7_+_39446247 | 0.09 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr19_-_29302249 | 0.09 |
ENSDART00000188751
|
srfbp1
|
serum response factor binding protein 1 |
| chr4_+_53145525 | 0.09 |
ENSDART00000166960
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr23_+_17437554 | 0.09 |
ENSDART00000184282
|
BX649300.3
|
|
| chr24_+_19863246 | 0.09 |
ENSDART00000165242
|
CU638714.1
|
|
| chr18_-_34170918 | 0.09 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr4_-_75982054 | 0.09 |
ENSDART00000168369
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr19_+_9186175 | 0.09 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr5_-_69425224 | 0.09 |
ENSDART00000158096
|
FQ311924.1
|
|
| chr8_+_53051701 | 0.09 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr21_+_15435946 | 0.08 |
ENSDART00000137491
|
si:dkey-11o15.10
|
si:dkey-11o15.10 |
| chr8_+_31435452 | 0.08 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr21_+_18877130 | 0.08 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr6_-_41135215 | 0.08 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr25_-_16600811 | 0.08 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr1_-_49950643 | 0.08 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr6_-_31987940 | 0.08 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_-_61592417 | 0.08 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr10_-_40964739 | 0.08 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr1_+_55643198 | 0.08 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr14_-_36763302 | 0.07 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr21_+_39948300 | 0.07 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr23_-_26227805 | 0.07 |
ENSDART00000158082
|
BX927204.1
|
|
| chr6_+_27361978 | 0.07 |
ENSDART00000065238
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr7_-_27038488 | 0.07 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr23_+_7518294 | 0.07 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr3_+_57788761 | 0.07 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr16_+_46725087 | 0.07 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3.3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.2 | 0.5 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.1 | 0.6 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.2 | GO:0097094 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.6 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 0.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.5 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |