Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
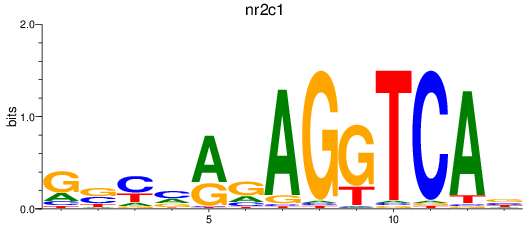
Results for nr2c1
Z-value: 1.09
Transcription factors associated with nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c1
|
ENSDARG00000045527 | nuclear receptor subfamily 2, group C, member 1 |
|
nr2c1
|
ENSDARG00000111803 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c1 | dr11_v1_chr4_-_25858620_25858620 | 0.95 | 1.5e-02 | Click! |
Activity profile of nr2c1 motif
Sorted Z-values of nr2c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_58353661 | 1.11 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr10_+_20128267 | 0.80 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr10_+_42374770 | 0.71 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr10_+_42374957 | 0.70 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr8_-_12468744 | 0.68 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr11_+_25101220 | 0.51 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr5_-_26330313 | 0.50 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr25_+_12012 | 0.47 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr17_+_53424415 | 0.47 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr24_-_1021318 | 0.46 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr1_+_58182400 | 0.44 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr12_+_19138452 | 0.44 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr5_+_29851433 | 0.43 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr23_-_45504991 | 0.42 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr3_+_24190207 | 0.42 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr17_-_49995366 | 0.42 |
ENSDART00000075223
|
cox7a2a
|
cytochrome c oxidase subunit VIIa polypeptide 2a (liver) |
| chr2_-_56649883 | 0.41 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr6_+_40523370 | 0.41 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr5_-_23280098 | 0.40 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr13_-_37127970 | 0.40 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_+_30002605 | 0.39 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr7_-_42206720 | 0.39 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr10_+_35417099 | 0.38 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr20_+_36806398 | 0.37 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr21_+_1119046 | 0.37 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr17_-_47305034 | 0.37 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr7_+_73332486 | 0.37 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr22_-_15280638 | 0.35 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr12_+_48681601 | 0.35 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr25_-_19420949 | 0.35 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr14_+_989733 | 0.34 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr13_+_713941 | 0.34 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr18_+_43183749 | 0.34 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr3_+_27770110 | 0.34 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr13_+_28819768 | 0.34 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr4_-_2975461 | 0.33 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_+_37612672 | 0.33 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_-_36020005 | 0.33 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr1_-_51720633 | 0.32 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr19_-_5369486 | 0.32 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr6_+_8176486 | 0.31 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr7_-_20756013 | 0.31 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr8_-_979735 | 0.30 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr16_-_2558653 | 0.30 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr6_-_39893501 | 0.30 |
ENSDART00000141611
ENSDART00000135631 ENSDART00000077662 ENSDART00000130613 |
myl6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr3_+_40289418 | 0.29 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr20_-_25877793 | 0.29 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr24_-_21674950 | 0.27 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr1_-_17650223 | 0.27 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr23_+_216012 | 0.26 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr15_-_28618502 | 0.26 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr1_-_19215336 | 0.26 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr22_-_38543630 | 0.26 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr18_+_1615 | 0.26 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr3_+_1179601 | 0.25 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr24_+_39137001 | 0.25 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_+_3056450 | 0.25 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr7_+_42206847 | 0.25 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr3_+_26035550 | 0.25 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr5_-_68927728 | 0.25 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr8_+_36948256 | 0.25 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr12_-_26423439 | 0.24 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr16_+_25809235 | 0.24 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr23_+_39346930 | 0.24 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr13_+_13805385 | 0.24 |
ENSDART00000166859
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr10_+_15340768 | 0.24 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr12_+_49125510 | 0.24 |
ENSDART00000185804
|
FO704607.1
|
|
| chr11_+_24819324 | 0.24 |
ENSDART00000184549
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr21_-_1972236 | 0.23 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr13_-_36566260 | 0.23 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr21_+_38089036 | 0.22 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr15_+_28119284 | 0.22 |
ENSDART00000188366
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr1_+_58221372 | 0.22 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr2_-_21618629 | 0.22 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr15_-_1844048 | 0.22 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_-_8197049 | 0.22 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr23_-_39636195 | 0.22 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr9_-_14504834 | 0.21 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr23_+_39346774 | 0.21 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr22_-_3595439 | 0.21 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr6_-_40583253 | 0.21 |
ENSDART00000032603
|
tspo
|
translocator protein |
| chr2_-_6548838 | 0.20 |
ENSDART00000052419
|
rgs18
|
regulator of G protein signaling 18 |
| chr12_+_26670778 | 0.20 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr5_-_10768258 | 0.20 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr3_-_33494163 | 0.20 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr11_+_36989696 | 0.19 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr1_-_53750522 | 0.19 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr13_+_45980163 | 0.19 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr20_+_53278746 | 0.19 |
ENSDART00000074249
|
gpr6
|
G protein-coupled receptor 6 |
| chr18_+_27515640 | 0.19 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr3_+_18062871 | 0.18 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr13_+_7241170 | 0.18 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr9_+_34433053 | 0.18 |
ENSDART00000137416
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr22_-_15587360 | 0.18 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr3_+_54047342 | 0.18 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr19_+_2631565 | 0.18 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr22_-_7129631 | 0.17 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr1_+_58303892 | 0.17 |
ENSDART00000147678
|
CR769768.1
|
|
| chr10_-_43404027 | 0.17 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr8_-_11146924 | 0.17 |
ENSDART00000126824
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr12_+_23866368 | 0.16 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr21_-_45728069 | 0.16 |
ENSDART00000185101
|
CU302316.1
|
|
| chr24_+_39641991 | 0.16 |
ENSDART00000142182
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr6_+_60036767 | 0.16 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr5_+_24156170 | 0.16 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr11_+_24819624 | 0.16 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr22_+_17399124 | 0.15 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr24_-_29586082 | 0.15 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr6_-_42111937 | 0.15 |
ENSDART00000181772
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr6_-_42112191 | 0.15 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr14_-_27121854 | 0.15 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr11_+_30729745 | 0.15 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_-_30609659 | 0.15 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr10_-_41400049 | 0.14 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr15_+_1004680 | 0.14 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr3_-_60308937 | 0.14 |
ENSDART00000136816
|
si:ch211-214b16.3
|
si:ch211-214b16.3 |
| chr19_-_24136233 | 0.14 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr15_+_24644016 | 0.14 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr1_+_58260886 | 0.14 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr8_-_11146687 | 0.13 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr17_+_1514711 | 0.13 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr1_-_44581937 | 0.13 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr9_+_24936496 | 0.13 |
ENSDART00000157474
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_59328030 | 0.13 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr7_+_50109239 | 0.13 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr15_+_24644251 | 0.13 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr23_-_46126444 | 0.13 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr23_-_2448234 | 0.12 |
ENSDART00000082097
|
LO018513.1
|
|
| chr6_-_51541488 | 0.12 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr18_-_158541 | 0.12 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_+_58119117 | 0.11 |
ENSDART00000146788
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr21_+_15883546 | 0.11 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr20_-_874807 | 0.11 |
ENSDART00000020506
|
snx14
|
sorting nexin 14 |
| chr7_-_26457208 | 0.11 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr1_-_43782 | 0.11 |
ENSDART00000168660
|
tmem39a
|
transmembrane protein 39A |
| chr23_-_35069805 | 0.11 |
ENSDART00000087219
|
BX294434.1
|
|
| chr12_-_15620090 | 0.10 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr7_+_7495034 | 0.10 |
ENSDART00000102629
ENSDART00000180475 |
nit1
|
nitrilase 1 |
| chr8_+_2530065 | 0.10 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr2_+_43755274 | 0.10 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr18_+_35742838 | 0.10 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr11_+_77526 | 0.10 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr7_-_69983948 | 0.10 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr1_+_58150000 | 0.09 |
ENSDART00000137836
|
si:ch211-15j1.3
|
si:ch211-15j1.3 |
| chr1_+_58442694 | 0.09 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr1_+_44582369 | 0.09 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr10_-_38443312 | 0.09 |
ENSDART00000141260
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr5_-_22615087 | 0.09 |
ENSDART00000146035
|
zgc:113208
|
zgc:113208 |
| chr3_+_30257582 | 0.09 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr3_+_18424244 | 0.08 |
ENSDART00000127796
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr16_-_38518270 | 0.08 |
ENSDART00000131899
|
rspo2
|
R-spondin 2 |
| chr8_+_10862353 | 0.08 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr18_+_38807239 | 0.08 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr10_+_29266392 | 0.08 |
ENSDART00000153577
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr20_-_26937453 | 0.08 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr15_+_47386939 | 0.07 |
ENSDART00000128224
|
FO904873.1
|
|
| chr1_+_58067815 | 0.07 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr16_-_560574 | 0.07 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr22_+_2417105 | 0.07 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr16_+_26012569 | 0.07 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr15_-_31213119 | 0.07 |
ENSDART00000182009
|
cuedc1b
|
CUE domain containing 1b |
| chr22_-_28373698 | 0.07 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr1_-_58868306 | 0.06 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr1_-_46859398 | 0.06 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr6_-_18393476 | 0.06 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr1_-_46343999 | 0.06 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr10_+_589501 | 0.06 |
ENSDART00000188415
|
LO018557.1
|
|
| chr25_+_19008497 | 0.05 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr8_+_8699085 | 0.05 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr24_-_31942656 | 0.05 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr23_-_45705525 | 0.05 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr15_+_21711671 | 0.05 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr19_+_20274944 | 0.04 |
ENSDART00000151237
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr13_-_31878043 | 0.04 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr7_-_46744080 | 0.04 |
ENSDART00000193328
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr25_+_22017182 | 0.04 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr8_+_8893535 | 0.04 |
ENSDART00000139150
|
otud5a
|
OTU deubiquitinase 5a |
| chr19_+_19652439 | 0.04 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr23_+_27912079 | 0.03 |
ENSDART00000171859
|
BX539336.1
|
|
| chr3_+_62216727 | 0.03 |
ENSDART00000180069
|
BX470259.4
|
|
| chr11_+_45343245 | 0.03 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr3_-_34561624 | 0.03 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr12_-_47899497 | 0.02 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr3_-_36419641 | 0.02 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr13_+_42406883 | 0.02 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_-_30553588 | 0.02 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr12_-_22509069 | 0.02 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr10_-_7555660 | 0.02 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr2_-_38141997 | 0.02 |
ENSDART00000143469
|
trim110
|
tripartite motif containing 110 |
| chr10_-_7555326 | 0.01 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr25_-_27665978 | 0.01 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr10_-_40939706 | 0.01 |
ENSDART00000059795
ENSDART00000190510 |
bmp1b
|
bone morphogenetic protein 1b |
| chr23_+_45611980 | 0.01 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr11_-_17713987 | 0.01 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr3_+_30968883 | 0.01 |
ENSDART00000085736
|
prf1.9
|
perforin 1.9 |
| chr11_+_3499111 | 0.01 |
ENSDART00000113096
ENSDART00000171621 |
pusl1
|
pseudouridylate synthase-like 1 |
| chr23_+_33963619 | 0.00 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr11_+_36243774 | 0.00 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr6_+_40952031 | 0.00 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr8_+_1433647 | 0.00 |
ENSDART00000160797
|
efna5b
|
ephrin-A5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0033335 | anal fin development(GO:0033335) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:0005689 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.3 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |