Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
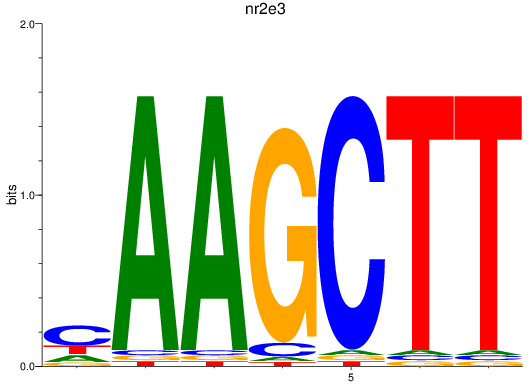
Results for nr2e3
Z-value: 0.85
Transcription factors associated with nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e3
|
ENSDARG00000045904 | nuclear receptor subfamily 2, group E, member 3 |
|
nr2e3
|
ENSDARG00000113178 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e3 | dr11_v1_chr25_+_22274642_22274642 | 0.59 | 3.0e-01 | Click! |
Activity profile of nr2e3 motif
Sorted Z-values of nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47633438 | 0.72 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr13_+_27316934 | 0.69 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr3_-_34801041 | 0.60 |
ENSDART00000103043
|
nsfa
|
N-ethylmaleimide-sensitive factor a |
| chr6_+_38427357 | 0.54 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_10275320 | 0.51 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr2_+_47582681 | 0.48 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr3_-_28665291 | 0.48 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr11_-_1024483 | 0.45 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr2_+_47582488 | 0.44 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr24_-_29997145 | 0.44 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr24_+_39158283 | 0.42 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr8_+_3820134 | 0.42 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr13_+_38520927 | 0.42 |
ENSDART00000137299
ENSDART00000111080 |
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr23_-_29502287 | 0.39 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr20_-_28698172 | 0.37 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr21_+_25643880 | 0.37 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr5_-_30588539 | 0.37 |
ENSDART00000086686
ENSDART00000159163 ENSDART00000142958 |
c2cd2l
|
c2cd2-like |
| chr4_+_4816695 | 0.37 |
ENSDART00000136962
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr5_+_58397646 | 0.35 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr8_+_16025554 | 0.35 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr10_+_7182168 | 0.34 |
ENSDART00000172766
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr3_-_27868183 | 0.33 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr16_-_12914288 | 0.33 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr8_-_23416362 | 0.33 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr23_+_19564392 | 0.33 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr14_-_43000836 | 0.32 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr6_+_55277419 | 0.32 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr14_-_21618005 | 0.32 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr5_-_28606916 | 0.31 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr22_+_34701848 | 0.31 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr20_+_34913069 | 0.31 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr2_+_34767171 | 0.31 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr12_+_9320783 | 0.31 |
ENSDART00000152526
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr13_+_27316632 | 0.30 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_+_13418812 | 0.30 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr3_+_7617353 | 0.30 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr25_+_21829777 | 0.30 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr8_+_31941650 | 0.29 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr5_+_36768674 | 0.29 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr22_-_3595439 | 0.29 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr20_-_31905968 | 0.29 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr10_-_7974155 | 0.29 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr3_+_15271943 | 0.29 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr24_+_2519761 | 0.29 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr14_-_46617228 | 0.28 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr6_+_38427570 | 0.28 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_37734973 | 0.28 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr10_+_34685135 | 0.28 |
ENSDART00000184999
|
nbeaa
|
neurobeachin a |
| chr20_-_44576949 | 0.27 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr19_+_37925616 | 0.27 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr3_-_43821381 | 0.27 |
ENSDART00000166021
|
snn
|
stannin |
| chr15_-_34213898 | 0.27 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr25_+_7670683 | 0.26 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr6_-_29143463 | 0.26 |
ENSDART00000087440
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr9_-_31596016 | 0.26 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr6_+_38381957 | 0.26 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr9_-_35334642 | 0.26 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr14_-_2202652 | 0.26 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr4_-_5019113 | 0.26 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr10_+_7182423 | 0.25 |
ENSDART00000186788
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr2_+_50608099 | 0.25 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr14_-_2163454 | 0.25 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr21_+_10076203 | 0.25 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr5_+_23242370 | 0.25 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr9_+_17348745 | 0.25 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr14_-_25599002 | 0.24 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr6_-_52400896 | 0.24 |
ENSDART00000187624
|
mmp24
|
matrix metallopeptidase 24 |
| chr14_-_46213571 | 0.23 |
ENSDART00000056734
|
setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_+_26632448 | 0.23 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_+_50448478 | 0.23 |
ENSDART00000098815
|
serinc4
|
serine incorporator 4 |
| chr8_+_29856013 | 0.23 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr14_-_30724165 | 0.23 |
ENSDART00000020936
|
fibpa
|
fibroblast growth factor (acidic) intracellular binding protein a |
| chr25_-_7686201 | 0.23 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr24_-_24163201 | 0.23 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr1_+_22851745 | 0.23 |
ENSDART00000138235
ENSDART00000016488 |
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr10_+_45128375 | 0.22 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr9_+_42063906 | 0.22 |
ENSDART00000048893
|
pcbp3
|
poly(rC) binding protein 3 |
| chr24_-_18809433 | 0.22 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr16_+_5774977 | 0.22 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr19_+_42231431 | 0.21 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr2_-_37043540 | 0.21 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_8017579 | 0.21 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr22_+_20208185 | 0.21 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr9_-_3671911 | 0.21 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr13_+_16522608 | 0.21 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr7_-_46781614 | 0.21 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr1_-_42778510 | 0.21 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr19_-_5812319 | 0.21 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr10_+_21776911 | 0.21 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr20_-_909953 | 0.21 |
ENSDART00000011283
|
cnr1
|
cannabinoid receptor 1 |
| chr18_-_7342950 | 0.21 |
ENSDART00000081356
|
caps2
|
calcyphosine 2 |
| chr10_+_24504464 | 0.21 |
ENSDART00000141387
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr12_+_42436328 | 0.21 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr9_-_35557397 | 0.20 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr8_-_14080534 | 0.20 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr3_+_17456428 | 0.20 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr17_-_3986236 | 0.20 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr16_-_13004166 | 0.20 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr12_-_10705916 | 0.20 |
ENSDART00000164038
|
FO704786.1
|
|
| chr24_+_15020402 | 0.20 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr24_+_15277216 | 0.20 |
ENSDART00000081449
|
socs6a
|
suppressor of cytokine signaling 6a |
| chr17_+_51499789 | 0.20 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr20_-_15132151 | 0.20 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr25_-_12203952 | 0.20 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr18_-_38087875 | 0.20 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr10_+_2981090 | 0.19 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr6_-_51386656 | 0.19 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr12_+_26471712 | 0.19 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr20_+_19238382 | 0.19 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr21_+_32332616 | 0.19 |
ENSDART00000154105
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr4_+_68476490 | 0.19 |
ENSDART00000168543
|
si:dkey-237g15.2
|
si:dkey-237g15.2 |
| chr22_-_17574511 | 0.19 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr3_-_43821191 | 0.19 |
ENSDART00000186816
|
snn
|
stannin |
| chr5_-_33607062 | 0.19 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr10_+_33902264 | 0.19 |
ENSDART00000193793
|
fryb
|
furry homolog b (Drosophila) |
| chr9_-_6661657 | 0.19 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr17_-_44247707 | 0.19 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr6_-_12314475 | 0.19 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr23_-_17657348 | 0.18 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr22_-_18022416 | 0.18 |
ENSDART00000141563
|
ncanb
|
neurocan b |
| chr9_+_30720048 | 0.18 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr9_-_12034444 | 0.18 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr1_-_54997746 | 0.18 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr1_+_32521469 | 0.18 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr7_-_35126374 | 0.18 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr9_+_6079528 | 0.18 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr20_-_34453623 | 0.18 |
ENSDART00000005609
|
kifap3a
|
kinesin-associated protein 3a |
| chr6_+_37754763 | 0.18 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr9_-_40935934 | 0.18 |
ENSDART00000155604
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr4_+_23223881 | 0.18 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr18_+_50276653 | 0.18 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr15_+_7992906 | 0.18 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr5_-_10946232 | 0.18 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr15_-_14486534 | 0.17 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr20_-_50239406 | 0.17 |
ENSDART00000128053
|
tmem121ab
|
transmembrane protein 121Ab |
| chr9_-_16133263 | 0.17 |
ENSDART00000077187
|
myo1b
|
myosin IB |
| chr18_-_8857137 | 0.17 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr23_-_28294763 | 0.17 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr18_-_38088099 | 0.17 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr6_-_35310224 | 0.17 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr11_-_37995501 | 0.17 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr5_-_43959972 | 0.17 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr12_+_25600685 | 0.17 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr22_+_31930650 | 0.17 |
ENSDART00000092090
|
dock3
|
dedicator of cytokinesis 3 |
| chr8_-_29851706 | 0.17 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr7_+_63325819 | 0.17 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr20_-_34868814 | 0.17 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr18_-_23875219 | 0.17 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr9_+_17983463 | 0.16 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr11_+_13071645 | 0.16 |
ENSDART00000162259
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr8_-_15398760 | 0.16 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr12_+_32368574 | 0.16 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr18_-_14734678 | 0.16 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr18_-_6824518 | 0.16 |
ENSDART00000141132
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr9_+_17982737 | 0.16 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr16_-_9383629 | 0.16 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr12_+_2631711 | 0.16 |
ENSDART00000114209
|
gdf10b
|
growth differentiation factor 10b |
| chr23_+_2778813 | 0.16 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr18_-_23874929 | 0.16 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_32338334 | 0.16 |
ENSDART00000065149
|
dock7
|
dedicator of cytokinesis 7 |
| chr10_+_15255198 | 0.16 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr5_+_32009080 | 0.16 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr10_+_15255012 | 0.16 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr18_-_7382003 | 0.16 |
ENSDART00000101238
ENSDART00000178284 |
bbs10
|
Bardet-Biedl syndrome 10 |
| chr20_+_32523576 | 0.16 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr6_-_13408680 | 0.16 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr14_-_2036604 | 0.16 |
ENSDART00000192446
|
BX005294.2
|
|
| chr4_-_63923523 | 0.16 |
ENSDART00000144330
|
si:dkey-179k24.1
|
si:dkey-179k24.1 |
| chr7_-_5487593 | 0.15 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr14_+_45028062 | 0.15 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr4_-_5077158 | 0.15 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr25_-_7925019 | 0.15 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr20_-_28349144 | 0.15 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr12_+_31744217 | 0.15 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr25_+_33033633 | 0.15 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr1_-_36151377 | 0.15 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr11_-_28911172 | 0.15 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr18_+_37120096 | 0.15 |
ENSDART00000147385
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr10_-_7909839 | 0.15 |
ENSDART00000139868
|
slc35e4
|
solute carrier family 35, member E4 |
| chr10_+_22782522 | 0.15 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chrM_+_8894 | 0.15 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr8_+_13364950 | 0.15 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr21_+_34976600 | 0.15 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr6_+_39222598 | 0.15 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr20_-_31252809 | 0.14 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr5_+_38263240 | 0.14 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr12_+_9320446 | 0.14 |
ENSDART00000112507
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr5_+_19712011 | 0.14 |
ENSDART00000131924
|
fam222a
|
family with sequence similarity 222, member A |
| chr7_+_21752168 | 0.14 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr24_+_37080771 | 0.14 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr21_+_10794914 | 0.14 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr8_-_1838315 | 0.14 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr6_-_46403475 | 0.14 |
ENSDART00000154148
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr22_+_1049367 | 0.14 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr20_+_29690901 | 0.14 |
ENSDART00000142669
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr21_-_7691293 | 0.14 |
ENSDART00000093339
|
pde8b
|
phosphodiesterase 8B |
| chr9_-_4506819 | 0.14 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr9_-_25444091 | 0.14 |
ENSDART00000002428
|
acvr2aa
|
activin A receptor type 2Aa |
| chr3_+_46425262 | 0.13 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr18_+_40355408 | 0.13 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048913 | Golgi vesicle docking(GO:0048211) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 0.3 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.1 | 0.3 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.3 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.3 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.1 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 1.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) multi-organism behavior(GO:0051705) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.0 | GO:0060137 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.0 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.4 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 1.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |