Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
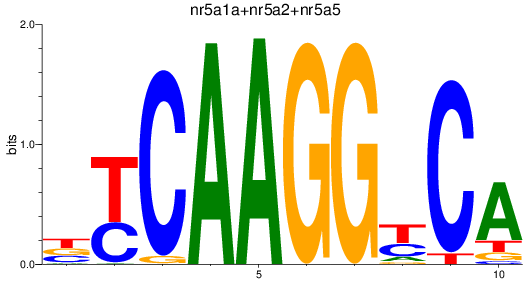
Results for nr5a1a+nr5a2+nr5a5
Z-value: 0.50
Transcription factors associated with nr5a1a+nr5a2+nr5a5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr5a5
|
ENSDARG00000039116 | nuclear receptor subfamily 5, group A, member 5 |
|
nr5a2
|
ENSDARG00000100940 | nuclear receptor subfamily 5, group A, member 2 |
|
nr5a1a
|
ENSDARG00000103176 | nuclear receptor subfamily 5, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr5a2 | dr11_v1_chr22_-_22719440_22719440 | -0.66 | 2.2e-01 | Click! |
| nr5a5 | dr11_v1_chr3_-_53465223_53465249 | -0.24 | 7.0e-01 | Click! |
| nr5a1a | dr11_v1_chr8_-_52871056_52871056 | 0.23 | 7.1e-01 | Click! |
Activity profile of nr5a1a+nr5a2+nr5a5 motif
Sorted Z-values of nr5a1a+nr5a2+nr5a5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24289641 | 0.88 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr13_-_2215213 | 0.38 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr19_-_30404096 | 0.37 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr23_-_5683147 | 0.37 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr8_-_29719393 | 0.37 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr13_+_50375800 | 0.36 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr11_-_25213651 | 0.36 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr23_+_18722915 | 0.36 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr15_+_20403903 | 0.34 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr23_+_18722715 | 0.34 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr6_-_15653494 | 0.29 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr19_-_30403922 | 0.29 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr3_+_39568290 | 0.29 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr22_+_7439476 | 0.28 |
ENSDART00000021594
ENSDART00000063389 |
zgc:92041
|
zgc:92041 |
| chr5_+_51594209 | 0.27 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr11_+_11230121 | 0.27 |
ENSDART00000172438
|
myom2a
|
myomesin 2a |
| chr16_-_17197546 | 0.27 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_+_22775253 | 0.27 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr6_-_35439406 | 0.26 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr7_+_48761646 | 0.26 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr3_+_24207243 | 0.25 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr6_-_29195642 | 0.25 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr22_+_5478353 | 0.24 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr9_+_24095677 | 0.23 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr8_-_21372446 | 0.23 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr17_-_32370047 | 0.22 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr21_-_22669669 | 0.21 |
ENSDART00000101784
|
gig2j
|
grass carp reovirus (GCRV)-induced gene 2j |
| chr25_-_13188678 | 0.20 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr21_-_17956739 | 0.20 |
ENSDART00000148154
|
stx2a
|
syntaxin 2a |
| chr4_+_2620751 | 0.20 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr14_-_29826659 | 0.19 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr16_+_31542645 | 0.19 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr1_+_9994811 | 0.19 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr3_-_34561624 | 0.19 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr1_-_8020589 | 0.19 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr16_-_29702447 | 0.17 |
ENSDART00000150617
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr19_-_32487469 | 0.17 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr17_+_5976683 | 0.17 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr23_+_19790962 | 0.17 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr18_-_26101800 | 0.17 |
ENSDART00000004692
|
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr7_-_20464468 | 0.17 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr2_-_22152797 | 0.17 |
ENSDART00000145188
|
cyp7a1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr6_-_13206255 | 0.17 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr12_-_48477031 | 0.16 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr14_-_12253309 | 0.16 |
ENSDART00000115101
|
myot
|
myotilin |
| chr20_+_40237441 | 0.16 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr3_+_13624815 | 0.16 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr6_-_1514767 | 0.16 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr22_+_7439186 | 0.16 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr15_+_404891 | 0.16 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr11_+_705727 | 0.16 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr21_-_17956416 | 0.15 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr1_+_29862074 | 0.15 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr22_+_7480465 | 0.15 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr21_-_5879897 | 0.15 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr1_+_50987535 | 0.15 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr19_+_18799319 | 0.15 |
ENSDART00000171843
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chrM_+_9735 | 0.15 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr14_+_45732081 | 0.14 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr14_-_21064199 | 0.14 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr22_+_661711 | 0.14 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr16_-_42013858 | 0.14 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr2_+_45191049 | 0.14 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr5_-_26566435 | 0.14 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr8_+_23213320 | 0.14 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr5_+_6672870 | 0.14 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr6_+_13206516 | 0.14 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr7_+_48555626 | 0.13 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr23_+_25708787 | 0.13 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr1_+_55662491 | 0.13 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr10_-_16065185 | 0.13 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr23_-_24488696 | 0.13 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr6_-_8480815 | 0.13 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr9_-_1970071 | 0.13 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr25_-_13408760 | 0.13 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr25_+_3104959 | 0.12 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr14_-_32884138 | 0.12 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr1_-_44940830 | 0.12 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr3_-_34060153 | 0.12 |
ENSDART00000151610
|
ighv1-1
|
immunoglobulin heavy variable 1-1 |
| chr3_-_32859335 | 0.12 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr5_-_33959868 | 0.12 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr14_-_36412473 | 0.12 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr20_+_37794633 | 0.12 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr1_+_52735484 | 0.11 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr4_-_77680276 | 0.11 |
ENSDART00000153262
|
si:ch211-250m6.2
|
si:ch211-250m6.2 |
| chr7_+_22293894 | 0.11 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr24_+_24809877 | 0.11 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr9_-_23891102 | 0.11 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr9_+_30585552 | 0.11 |
ENSDART00000160590
ENSDART00000189815 ENSDART00000143766 |
tbc1d4
|
TBC1 domain family, member 4 |
| chr25_+_25124684 | 0.11 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr22_+_344763 | 0.11 |
ENSDART00000181934
|
CU914780.1
|
|
| chr22_+_661505 | 0.11 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr2_+_36620011 | 0.11 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr9_-_34507680 | 0.11 |
ENSDART00000113656
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr18_-_21188768 | 0.11 |
ENSDART00000060166
|
got2a
|
glutamic-oxaloacetic transaminase 2a, mitochondrial |
| chr6_-_48082525 | 0.11 |
ENSDART00000192049
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr25_-_13202208 | 0.10 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr16_-_25085327 | 0.10 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr7_-_38658411 | 0.10 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr6_+_13207139 | 0.10 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr14_+_6159162 | 0.10 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr1_-_14533611 | 0.10 |
ENSDART00000180678
|
CR847936.6
|
|
| chr14_-_5678457 | 0.10 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr5_+_27429872 | 0.10 |
ENSDART00000087894
|
zgc:165555
|
zgc:165555 |
| chr19_+_24488403 | 0.10 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr21_+_198269 | 0.10 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr15_-_4485828 | 0.10 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr20_-_52967878 | 0.10 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr14_+_15484544 | 0.10 |
ENSDART00000188649
|
CR382326.1
|
|
| chr16_-_5721386 | 0.09 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr22_+_19266995 | 0.09 |
ENSDART00000133995
ENSDART00000144963 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr25_-_3230187 | 0.09 |
ENSDART00000160545
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr1_+_580642 | 0.09 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr15_-_2841677 | 0.09 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr11_+_807153 | 0.09 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr22_-_36876133 | 0.09 |
ENSDART00000147006
|
kng1
|
kininogen 1 |
| chr16_+_29509133 | 0.09 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr25_-_13050959 | 0.09 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr17_+_37227936 | 0.09 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr14_+_35892802 | 0.09 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr9_+_15893093 | 0.09 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr18_+_48423973 | 0.09 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr7_-_33351485 | 0.09 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr2_+_45552901 | 0.09 |
ENSDART00000131701
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr7_+_45400716 | 0.08 |
ENSDART00000006868
|
uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr12_+_43585527 | 0.08 |
ENSDART00000161590
|
clrn3
|
clarin 3 |
| chr3_-_32275975 | 0.08 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr3_+_1182315 | 0.08 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr14_+_20941534 | 0.08 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr4_+_72797711 | 0.08 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr7_+_20467549 | 0.08 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr21_+_26071874 | 0.08 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr6_+_22337081 | 0.08 |
ENSDART00000128047
ENSDART00000138930 |
uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr10_-_22106331 | 0.08 |
ENSDART00000187654
|
BX005027.2
|
|
| chr17_-_8862424 | 0.08 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr3_-_34136368 | 0.08 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr24_+_29912509 | 0.08 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr14_+_6159356 | 0.08 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr6_+_18142623 | 0.08 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr21_+_5882300 | 0.08 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr9_-_44953664 | 0.08 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr2_-_5199431 | 0.08 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr12_-_25201576 | 0.08 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr2_-_14656669 | 0.08 |
ENSDART00000190219
|
CABZ01026681.1
|
|
| chr7_+_30178253 | 0.08 |
ENSDART00000002075
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr21_+_5080789 | 0.08 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr7_+_34688527 | 0.08 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr1_-_43983405 | 0.08 |
ENSDART00000148883
|
si:dkey-22i16.9
|
si:dkey-22i16.9 |
| chr19_+_7864767 | 0.08 |
ENSDART00000137540
ENSDART00000151642 |
si:dkeyp-85e10.3
|
si:dkeyp-85e10.3 |
| chr17_-_26610814 | 0.08 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr13_+_42124566 | 0.08 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_-_30583668 | 0.07 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr23_-_16737161 | 0.07 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr7_-_59256806 | 0.07 |
ENSDART00000167955
|
m1ap
|
meiosis 1 associated protein |
| chr2_-_42091926 | 0.07 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr14_-_35414559 | 0.07 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr7_-_61901695 | 0.07 |
ENSDART00000110328
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr20_+_30939178 | 0.07 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr1_+_36772348 | 0.07 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr22_+_21252790 | 0.07 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr8_+_24740013 | 0.07 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr14_-_35892767 | 0.07 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr10_+_9092815 | 0.07 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr24_+_26432541 | 0.07 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr22_+_30046365 | 0.07 |
ENSDART00000192546
|
add3a
|
adducin 3 (gamma) a |
| chr9_+_27354414 | 0.07 |
ENSDART00000163804
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr1_-_55118745 | 0.07 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr9_+_27371240 | 0.07 |
ENSDART00000060337
|
tlr20.3
|
toll-like receptor 20, tandem duplicate 3 |
| chr9_+_33145522 | 0.07 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr7_-_58843193 | 0.07 |
ENSDART00000167231
|
mrpl15
|
mitochondrial ribosomal protein L15 |
| chr22_+_7742211 | 0.07 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr17_-_51202339 | 0.07 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr3_-_34054081 | 0.07 |
ENSDART00000151590
|
ighv1-2
|
immunoglobulin heavy variable 1-2 |
| chr4_-_16345227 | 0.07 |
ENSDART00000079521
|
kera
|
keratocan |
| chr22_+_6293563 | 0.07 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr6_+_8176486 | 0.07 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr18_-_6803424 | 0.07 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr3_+_2669813 | 0.07 |
ENSDART00000014205
|
CR388047.1
|
|
| chr6_+_9898683 | 0.07 |
ENSDART00000005573
|
tmem237b
|
transmembrane protein 237b |
| chr3_-_60288533 | 0.07 |
ENSDART00000181981
ENSDART00000193313 |
si:ch211-214b16.3
|
si:ch211-214b16.3 |
| chr9_-_44953349 | 0.07 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr3_+_26081343 | 0.07 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr7_-_28696556 | 0.07 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr23_+_42338325 | 0.07 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr8_+_52530889 | 0.07 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr4_-_17812131 | 0.07 |
ENSDART00000025731
|
spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr3_-_1434135 | 0.07 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr22_-_36934040 | 0.06 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr25_-_26893006 | 0.06 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr5_+_30089715 | 0.06 |
ENSDART00000078114
|
timm8b
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr19_-_12145390 | 0.06 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr2_-_51511494 | 0.06 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr7_-_12821277 | 0.06 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr3_-_40301467 | 0.06 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr12_+_7399403 | 0.06 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr9_+_27348809 | 0.06 |
ENSDART00000147540
|
tlr20.1
|
toll-like receptor 20, tandem duplicate 1 |
| chr22_+_7462997 | 0.06 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr6_+_36381709 | 0.06 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr19_+_4051535 | 0.06 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr14_+_3056713 | 0.06 |
ENSDART00000165745
|
si:dkey-201i24.3
|
si:dkey-201i24.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr5a1a+nr5a2+nr5a5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.3 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.3 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.2 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:2000379 | positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.1 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.1 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.9 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |