Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
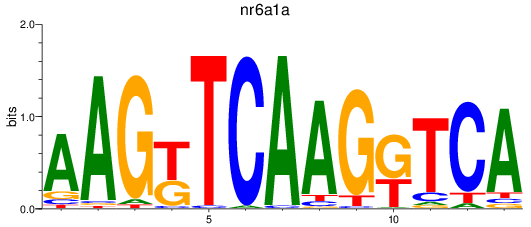
Results for nr6a1a
Z-value: 2.37
Transcription factors associated with nr6a1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1a
|
ENSDARG00000101508 | nuclear receptor subfamily 6, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1a | dr11_v1_chr8_-_52938439_52938439 | -0.73 | 1.6e-01 | Click! |
Activity profile of nr6a1a motif
Sorted Z-values of nr6a1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_12660318 | 3.98 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr20_+_10539293 | 3.03 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr4_-_17409533 | 2.13 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr22_-_24858042 | 2.08 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr3_+_19299309 | 2.01 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr20_+_10545514 | 2.01 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr8_+_1769475 | 1.82 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr15_-_26549693 | 1.67 |
ENSDART00000186432
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_8498908 | 1.52 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr6_-_8498676 | 1.46 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr9_-_48700806 | 1.42 |
ENSDART00000026210
|
rdh1
|
retinol dehydrogenase 1 |
| chr20_-_30370884 | 1.40 |
ENSDART00000062429
|
allc
|
allantoicase |
| chr2_-_50225411 | 1.32 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr3_-_3448095 | 1.28 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr15_+_14854666 | 1.27 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr16_-_45235947 | 1.27 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr5_-_37103487 | 1.26 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr21_-_30658509 | 1.24 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr7_+_19482084 | 1.19 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr16_+_49249669 | 1.19 |
ENSDART00000181899
|
LO018128.1
|
|
| chr21_-_43636595 | 1.17 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr25_-_29415369 | 1.16 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr2_+_51028269 | 1.14 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr8_+_19514294 | 1.13 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr3_+_21189766 | 1.12 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr19_-_14155781 | 1.07 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr24_-_9989634 | 1.06 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr9_-_48701035 | 1.06 |
ENSDART00000178332
|
rdh1
|
retinol dehydrogenase 1 |
| chr25_+_34576067 | 1.01 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr25_+_3327071 | 1.01 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr10_-_22497980 | 1.01 |
ENSDART00000185549
|
CT573337.2
|
|
| chr24_-_9979342 | 1.01 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr12_-_4346085 | 1.00 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr7_-_60831082 | 0.99 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr18_+_8340886 | 0.98 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr25_+_22107643 | 0.98 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr3_+_39540014 | 0.98 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr22_+_25086942 | 0.98 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr25_+_10830269 | 0.98 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr2_-_37210397 | 0.97 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr22_+_25086567 | 0.97 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr15_+_28410664 | 0.94 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr19_+_32855139 | 0.92 |
ENSDART00000052082
|
rpl30
|
ribosomal protein L30 |
| chr5_-_41531629 | 0.91 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr19_-_42571829 | 0.90 |
ENSDART00000102606
|
zgc:103438
|
zgc:103438 |
| chr25_+_3326885 | 0.89 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr8_-_14091886 | 0.86 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr22_-_23781083 | 0.86 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr16_+_29509133 | 0.83 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr1_-_7998493 | 0.83 |
ENSDART00000185901
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr23_-_10175898 | 0.83 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr13_+_8306399 | 0.81 |
ENSDART00000080339
|
galm
|
galactose mutarotase |
| chr20_+_25581627 | 0.81 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr11_+_14333441 | 0.80 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr4_+_77948517 | 0.79 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_-_23569833 | 0.78 |
ENSDART00000138889
|
si:dkey-58b18.10
|
si:dkey-58b18.10 |
| chr12_+_45676667 | 0.77 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr9_+_42270043 | 0.76 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr10_+_37927100 | 0.76 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr9_+_13682133 | 0.76 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr8_-_8489886 | 0.74 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr22_-_17781213 | 0.74 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr3_+_32112004 | 0.72 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr21_+_20901505 | 0.71 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr7_+_52122224 | 0.71 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr3_-_36823643 | 0.70 |
ENSDART00000131340
ENSDART00000185075 |
abcc6b.2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 2 |
| chr15_+_26600611 | 0.70 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr2_+_30032303 | 0.70 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr3_-_1970820 | 0.69 |
ENSDART00000136467
|
si:ch211-254c8.3
|
si:ch211-254c8.3 |
| chr22_-_17462895 | 0.68 |
ENSDART00000160415
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr5_-_41834999 | 0.68 |
ENSDART00000135772
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr7_-_37917517 | 0.67 |
ENSDART00000173795
|
heatr3
|
HEAT repeat containing 3 |
| chr5_-_50992690 | 0.67 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr3_+_2669813 | 0.67 |
ENSDART00000014205
|
CR388047.1
|
|
| chr3_-_10677890 | 0.67 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr9_-_14055959 | 0.67 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr22_+_21152847 | 0.66 |
ENSDART00000007321
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr2_-_23538794 | 0.66 |
ENSDART00000143857
|
si:dkey-58b18.10
|
si:dkey-58b18.10 |
| chr3_+_24612191 | 0.65 |
ENSDART00000190998
|
zgc:171506
|
zgc:171506 |
| chr10_+_33393829 | 0.65 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr24_+_19986432 | 0.65 |
ENSDART00000184402
|
CR381686.5
|
|
| chr25_+_18587338 | 0.65 |
ENSDART00000180287
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr8_-_2153147 | 0.64 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr22_-_22337382 | 0.64 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr10_-_36793412 | 0.64 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr25_-_13188678 | 0.64 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr21_-_17956739 | 0.63 |
ENSDART00000148154
|
stx2a
|
syntaxin 2a |
| chr1_-_11104805 | 0.63 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr9_-_1970071 | 0.63 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr2_-_20120904 | 0.62 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr2_-_23600821 | 0.62 |
ENSDART00000146217
|
BX677666.1
|
|
| chr3_-_7656059 | 0.62 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr9_-_12888082 | 0.61 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr21_-_5879897 | 0.61 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr19_+_7878122 | 0.61 |
ENSDART00000131917
|
si:dkeyp-85e10.2
|
si:dkeyp-85e10.2 |
| chr10_+_40629616 | 0.61 |
ENSDART00000147476
|
CR396590.9
|
|
| chr5_-_11971946 | 0.61 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr8_-_45759137 | 0.61 |
ENSDART00000123878
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr7_-_58826164 | 0.60 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chr22_+_21153031 | 0.60 |
ENSDART00000166342
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr3_+_13469704 | 0.60 |
ENSDART00000166806
|
zgc:77748
|
zgc:77748 |
| chr21_-_22869802 | 0.58 |
ENSDART00000189917
|
polq
|
polymerase (DNA directed), theta |
| chr5_+_9259971 | 0.58 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr19_+_4038589 | 0.57 |
ENSDART00000169271
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr21_+_43702016 | 0.57 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr19_-_5135345 | 0.56 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr25_-_13202208 | 0.56 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr17_+_450956 | 0.56 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr3_-_36824190 | 0.56 |
ENSDART00000172943
|
abcc6b.2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 2 |
| chr5_+_22579975 | 0.55 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr10_+_44719697 | 0.55 |
ENSDART00000158087
|
scarb1
|
scavenger receptor class B, member 1 |
| chr25_-_35143360 | 0.55 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr25_+_14092871 | 0.54 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr16_-_12319822 | 0.53 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr16_-_34195002 | 0.53 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr3_+_30921246 | 0.53 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr5_+_50879545 | 0.53 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr21_-_17956416 | 0.52 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr2_-_44344321 | 0.52 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr18_+_36631923 | 0.52 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr8_+_44703864 | 0.51 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr16_+_21790870 | 0.51 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr8_-_20245892 | 0.51 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr18_-_26854959 | 0.51 |
ENSDART00000057553
|
ch25hl1.1
|
cholesterol 25-hydroxylase like 1, tandem duplicate 1 |
| chr10_-_11383900 | 0.50 |
ENSDART00000101045
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr7_+_21272833 | 0.50 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr14_+_34514336 | 0.50 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr14_+_16034447 | 0.50 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr10_+_7719796 | 0.50 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr5_-_26936861 | 0.49 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr5_+_9434288 | 0.49 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr23_+_36087219 | 0.49 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr25_-_34512102 | 0.49 |
ENSDART00000087450
|
klf13
|
Kruppel-like factor 13 |
| chr16_+_23960933 | 0.48 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr3_+_31662126 | 0.48 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr3_-_32957702 | 0.48 |
ENSDART00000146586
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr8_+_42717005 | 0.48 |
ENSDART00000181656
ENSDART00000180320 ENSDART00000181293 |
zgc:194007
|
zgc:194007 |
| chr5_-_41841675 | 0.48 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr1_+_24557414 | 0.48 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_+_1749793 | 0.48 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr12_-_17024079 | 0.48 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr13_-_14926318 | 0.47 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr14_+_6535426 | 0.47 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr4_+_29620959 | 0.47 |
ENSDART00000133110
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr15_-_41720173 | 0.47 |
ENSDART00000155585
ENSDART00000075909 |
ftr76
|
finTRIM family, member 76 |
| chr8_+_44478294 | 0.46 |
ENSDART00000006898
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr5_+_1983495 | 0.46 |
ENSDART00000179860
|
CT573103.2
|
|
| chr24_+_34069675 | 0.46 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr3_+_4346854 | 0.46 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr12_+_30788912 | 0.45 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr2_-_42492201 | 0.45 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr24_-_25256230 | 0.45 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr11_+_2198831 | 0.45 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr23_-_43393083 | 0.45 |
ENSDART00000112758
|
zgc:174862
|
zgc:174862 |
| chr19_-_703898 | 0.45 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr7_-_24181159 | 0.43 |
ENSDART00000181206
|
zgc:153151
|
zgc:153151 |
| chr7_-_15370042 | 0.43 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr5_-_9090178 | 0.43 |
ENSDART00000091472
|
kcnv2b
|
potassium channel, subfamily V, member 2b |
| chr22_+_9126557 | 0.43 |
ENSDART00000190669
|
si:ch211-156p11.1
|
si:ch211-156p11.1 |
| chr6_+_42338309 | 0.43 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr3_+_23697997 | 0.43 |
ENSDART00000184299
ENSDART00000078466 |
hoxb3a
|
homeobox B3a |
| chr9_-_44939104 | 0.42 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr4_+_58668661 | 0.42 |
ENSDART00000188815
|
CR388148.2
|
|
| chr4_-_34775762 | 0.42 |
ENSDART00000167262
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr17_-_6765877 | 0.42 |
ENSDART00000193102
|
CR352340.1
|
|
| chr5_-_9948497 | 0.41 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr16_+_23961276 | 0.41 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr22_-_6229275 | 0.41 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr3_+_12484008 | 0.41 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr18_+_8912113 | 0.41 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr5_+_8246603 | 0.41 |
ENSDART00000167373
|
si:ch1073-475a24.1
|
si:ch1073-475a24.1 |
| chr9_+_29548195 | 0.40 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr25_+_23280220 | 0.40 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr2_+_52232630 | 0.40 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr14_+_32838110 | 0.40 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr21_-_33126697 | 0.40 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr1_+_56886214 | 0.40 |
ENSDART00000152718
ENSDART00000182408 |
si:ch211-1f22.8
|
si:ch211-1f22.8 |
| chr4_-_39479644 | 0.39 |
ENSDART00000183332
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr17_+_6538733 | 0.39 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr1_-_8566567 | 0.39 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr9_+_24088062 | 0.38 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr20_-_43771871 | 0.38 |
ENSDART00000153304
|
matn3a
|
matrilin 3a |
| chr4_-_75795899 | 0.38 |
ENSDART00000157925
|
si:dkey-261j11.1
|
si:dkey-261j11.1 |
| chr4_+_76973771 | 0.38 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr8_+_22472584 | 0.38 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr17_-_8862424 | 0.37 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr3_-_39627412 | 0.37 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr3_-_49015203 | 0.37 |
ENSDART00000186138
|
BX769173.2
|
|
| chr14_+_32837914 | 0.37 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr25_-_16742438 | 0.37 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr17_+_38566717 | 0.37 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr4_+_60547709 | 0.37 |
ENSDART00000158732
|
si:dkey-211i20.5
|
si:dkey-211i20.5 |
| chr25_-_12804450 | 0.37 |
ENSDART00000169717
|
ca5a
|
carbonic anhydrase Va |
| chr20_+_35445462 | 0.36 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr7_-_16034324 | 0.36 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr25_+_17860962 | 0.36 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr16_-_17197546 | 0.36 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_7818368 | 0.36 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr4_-_38829098 | 0.36 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr15_-_28082310 | 0.36 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.6 | 4.0 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.5 | 1.4 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.4 | 2.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.4 | 1.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 1.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 0.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.7 | GO:0000480 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.6 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 3.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.5 | GO:0035376 | sterol import(GO:0035376) recognition of apoptotic cell(GO:0043654) cholesterol import(GO:0070508) |
| 0.2 | 2.1 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.9 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.2 | 0.8 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.5 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.8 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.5 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.6 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 1.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.4 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.1 | 9.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.5 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.3 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.8 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.6 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.7 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.4 | GO:0016037 | light absorption(GO:0016037) |
| 0.1 | 0.2 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 0.3 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 1.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.6 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 2.1 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 1.3 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 1.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 1.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 1.0 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.1 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 2.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 1.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.5 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 14.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.8 | 4.0 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.5 | 2.0 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 3.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 2.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 1.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.3 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 2.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 9.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.4 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.5 | GO:0010853 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 1.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 2.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.7 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.7 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 1.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 2.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |