Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
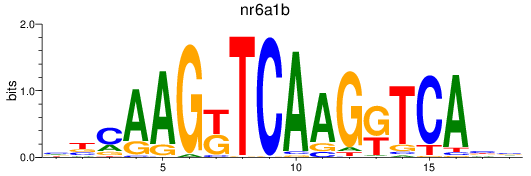
Results for nr6a1b
Z-value: 0.78
Transcription factors associated with nr6a1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1b
|
ENSDARG00000014480 | nuclear receptor subfamily 6, group A, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1b | dr11_v1_chr21_+_8198652_8198652 | -0.14 | 8.3e-01 | Click! |
Activity profile of nr6a1b motif
Sorted Z-values of nr6a1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10175898 | 1.02 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr21_+_11401247 | 0.61 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr7_-_15370042 | 0.52 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr19_+_7878122 | 0.50 |
ENSDART00000131917
|
si:dkeyp-85e10.2
|
si:dkeyp-85e10.2 |
| chr8_-_14091886 | 0.48 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr9_+_42270043 | 0.40 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr3_+_39540014 | 0.39 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr14_-_5678457 | 0.38 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr8_-_23578660 | 0.35 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr8_-_2153147 | 0.35 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr16_-_45235947 | 0.34 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr20_+_4392687 | 0.33 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr8_+_22472584 | 0.31 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr15_-_41720173 | 0.29 |
ENSDART00000155585
ENSDART00000075909 |
ftr76
|
finTRIM family, member 76 |
| chr12_-_17024079 | 0.29 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr15_+_19682013 | 0.28 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr8_+_22405477 | 0.26 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr16_+_49249669 | 0.26 |
ENSDART00000181899
|
LO018128.1
|
|
| chr3_+_1749793 | 0.25 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr11_+_45219558 | 0.25 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr4_+_9836465 | 0.25 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr10_+_29260096 | 0.24 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr13_+_8306399 | 0.21 |
ENSDART00000080339
|
galm
|
galactose mutarotase |
| chr4_+_9478500 | 0.21 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr10_+_37927100 | 0.20 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr20_+_10539293 | 0.20 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr4_+_69712223 | 0.19 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr3_+_32112004 | 0.18 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr12_-_30583668 | 0.18 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr4_-_34775762 | 0.17 |
ENSDART00000167262
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr3_-_7656059 | 0.17 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr5_-_50992690 | 0.16 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr1_-_55118745 | 0.16 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr17_-_53348486 | 0.15 |
ENSDART00000161183
|
CABZ01066926.1
|
|
| chr1_+_17527342 | 0.15 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr11_+_45287541 | 0.15 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr4_+_76973771 | 0.15 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr5_-_11971946 | 0.15 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr20_+_9128829 | 0.15 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr19_-_5135345 | 0.14 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr6_+_39506043 | 0.14 |
ENSDART00000086260
|
CR388364.1
|
|
| chr16_+_27224068 | 0.14 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr9_-_1970071 | 0.14 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr15_+_19681718 | 0.14 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr7_+_19482084 | 0.13 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr4_-_49582758 | 0.13 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr2_-_37210397 | 0.13 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr1_+_17527931 | 0.13 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr4_-_34776147 | 0.12 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr1_-_11104805 | 0.12 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr24_-_25256230 | 0.12 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr4_-_38829098 | 0.11 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr4_+_57192681 | 0.11 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_71267461 | 0.11 |
ENSDART00000161219
ENSDART00000151838 |
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr12_+_30788912 | 0.11 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr21_-_5879897 | 0.11 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr4_+_58668661 | 0.11 |
ENSDART00000188815
|
CR388148.2
|
|
| chr2_-_50225411 | 0.11 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr10_-_29120515 | 0.10 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
| chr17_+_18117029 | 0.10 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr13_-_22843562 | 0.10 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr17_+_18117358 | 0.10 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr1_-_49947290 | 0.10 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr22_-_17781213 | 0.09 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr1_-_49947482 | 0.08 |
ENSDART00000143398
|
sgms2
|
sphingomyelin synthase 2 |
| chr4_-_43616331 | 0.08 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_70263609 | 0.08 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr7_+_29461060 | 0.08 |
ENSDART00000075721
|
foxb1b
|
forkhead box B1b |
| chr22_+_24603930 | 0.08 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr4_+_34119824 | 0.08 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr8_+_25145464 | 0.07 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr4_-_63166758 | 0.07 |
ENSDART00000192765
|
CR450780.1
|
|
| chr4_-_9836342 | 0.07 |
ENSDART00000150519
|
ttc41
|
tetratricopeptide repeat domain 41 |
| chr15_+_11840311 | 0.07 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr23_-_545942 | 0.07 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr10_-_16868211 | 0.06 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr7_-_20453661 | 0.06 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr23_+_17102960 | 0.06 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr4_+_47782411 | 0.06 |
ENSDART00000161728
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr7_-_30639385 | 0.06 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr4_+_67994238 | 0.06 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr2_+_13694450 | 0.06 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr24_+_19542323 | 0.05 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr9_+_32301456 | 0.05 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr8_-_54216499 | 0.05 |
ENSDART00000193678
|
mbd4
|
methyl-CpG binding domain protein 4 |
| chr3_+_2669813 | 0.05 |
ENSDART00000014205
|
CR388047.1
|
|
| chr6_+_27418541 | 0.05 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr4_-_71470455 | 0.05 |
ENSDART00000172685
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr1_+_57348756 | 0.04 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr16_-_27224000 | 0.04 |
ENSDART00000126347
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr16_+_48616220 | 0.03 |
ENSDART00000004751
ENSDART00000130045 |
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr5_-_3839285 | 0.03 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr4_-_47095583 | 0.03 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr4_-_34480599 | 0.03 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr25_+_10830269 | 0.03 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr4_-_47096170 | 0.03 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr9_-_27749936 | 0.03 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr3_-_39627412 | 0.03 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr13_+_713941 | 0.03 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr4_-_63559311 | 0.03 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr23_-_19230627 | 0.02 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr9_-_7287375 | 0.02 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr4_-_44509477 | 0.02 |
ENSDART00000102949
ENSDART00000135262 |
si:dkeyp-100h4.1
|
si:dkeyp-100h4.1 |
| chr3_-_49015203 | 0.02 |
ENSDART00000186138
|
BX769173.2
|
|
| chr11_-_5563498 | 0.02 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr7_+_35268054 | 0.01 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr15_+_25156121 | 0.01 |
ENSDART00000189709
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr9_+_27371240 | 0.01 |
ENSDART00000060337
|
tlr20.3
|
toll-like receptor 20, tandem duplicate 3 |
| chr20_-_53435483 | 0.01 |
ENSDART00000135091
|
mettl24
|
methyltransferase like 24 |
| chr1_+_29862074 | 0.01 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr10_-_22057001 | 0.00 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr3_-_13599482 | 0.00 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.3 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.6 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0003272 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |