Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
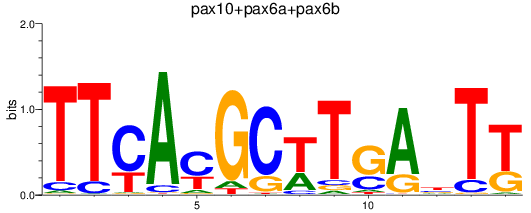
Results for pax10+pax6a+pax6b
Z-value: 0.85
Transcription factors associated with pax10+pax6a+pax6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax6b
|
ENSDARG00000045936 | paired box 6b |
|
pax10
|
ENSDARG00000053364 | paired box 10 |
|
pax6a
|
ENSDARG00000103379 | paired box 6a |
|
pax10
|
ENSDARG00000111379 | paired box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax10 | dr11_v1_chr3_+_32546499_32546499 | -0.86 | 6.1e-02 | Click! |
| pax6b | dr11_v1_chr7_+_15872357_15872357 | -0.82 | 9.1e-02 | Click! |
| pax6a | dr11_v1_chr25_-_15040369_15040442 | -0.76 | 1.3e-01 | Click! |
Activity profile of pax10+pax6a+pax6b motif
Sorted Z-values of pax10+pax6a+pax6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38306010 | 0.79 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr7_+_38750871 | 0.75 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr10_-_8053385 | 0.64 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_-_8046764 | 0.60 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr21_-_25756119 | 0.59 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr9_-_32847642 | 0.56 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr18_+_62932 | 0.53 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr2_+_11031360 | 0.47 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr25_+_22320738 | 0.45 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_-_8053753 | 0.44 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr13_-_20381485 | 0.43 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr14_-_1454045 | 0.41 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr23_+_44611864 | 0.40 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr23_+_44614056 | 0.36 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr11_+_11175814 | 0.35 |
ENSDART00000183864
|
CR847973.1
|
|
| chr7_-_20582842 | 0.35 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr12_+_4900981 | 0.35 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr8_-_8489685 | 0.32 |
ENSDART00000131849
ENSDART00000064113 |
abt1
|
activator of basal transcription 1 |
| chr23_+_17926279 | 0.32 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr21_-_2322102 | 0.31 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr6_+_23887314 | 0.31 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr8_+_52442622 | 0.30 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr8_-_8489886 | 0.30 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr25_-_31423493 | 0.30 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr25_-_27842654 | 0.30 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr21_+_27382893 | 0.29 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr1_-_54706039 | 0.29 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr25_-_4148719 | 0.29 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr5_+_57924611 | 0.27 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr13_-_10945288 | 0.27 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr15_-_12270857 | 0.26 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr2_+_6255434 | 0.26 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr7_+_59649746 | 0.26 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr3_+_43086548 | 0.25 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr19_+_30990815 | 0.25 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr9_+_8365398 | 0.24 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr20_+_2281933 | 0.24 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr22_-_5006801 | 0.24 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr19_-_15192840 | 0.24 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr19_-_11238620 | 0.24 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr4_-_5370468 | 0.23 |
ENSDART00000133423
ENSDART00000184963 |
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr9_-_1970071 | 0.23 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr2_+_21000334 | 0.23 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr16_-_13992646 | 0.22 |
ENSDART00000139623
|
si:dkey-85k15.6
|
si:dkey-85k15.6 |
| chr15_-_1745408 | 0.22 |
ENSDART00000182311
|
stx1a
|
syntaxin 1A (brain) |
| chr14_-_858985 | 0.22 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr17_+_8799661 | 0.21 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr13_-_22862133 | 0.21 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr18_+_45781115 | 0.21 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr8_+_37111643 | 0.21 |
ENSDART00000061336
|
ren
|
renin |
| chr18_+_45781516 | 0.21 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr11_+_24298432 | 0.21 |
ENSDART00000138487
|
si:dkey-76p14.2
|
si:dkey-76p14.2 |
| chr9_+_23003208 | 0.21 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr16_+_13993285 | 0.21 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr10_-_17501528 | 0.21 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr13_+_10945337 | 0.20 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr6_-_58757131 | 0.20 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr7_+_59649399 | 0.20 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr25_-_28926631 | 0.20 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr20_-_25486384 | 0.20 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr9_+_47966167 | 0.20 |
ENSDART00000183345
|
gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr16_+_11779761 | 0.20 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr25_-_37284370 | 0.20 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_41037541 | 0.20 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr12_+_25845494 | 0.20 |
ENSDART00000170582
|
glud1b
|
glutamate dehydrogenase 1b |
| chr12_-_35949936 | 0.20 |
ENSDART00000192583
|
AL954682.1
|
|
| chr15_-_14625373 | 0.20 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr15_+_9121054 | 0.19 |
ENSDART00000193889
|
ptgir
|
prostaglandin I2 receptor |
| chr5_-_57361594 | 0.19 |
ENSDART00000112793
|
cdc26
|
cell division cycle 26 homolog |
| chr1_+_55239160 | 0.19 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr17_-_12196865 | 0.19 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr19_-_4793263 | 0.18 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr18_-_14901437 | 0.18 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr18_-_27897217 | 0.18 |
ENSDART00000175259
|
iqcg
|
IQ motif containing G |
| chr13_-_39160018 | 0.18 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr17_+_8799451 | 0.18 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr23_-_19230627 | 0.18 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr24_-_38197040 | 0.17 |
ENSDART00000137949
ENSDART00000105639 |
igic1s1
|
immunoglobulin light iota constant 1, s1 |
| chr5_+_22393501 | 0.17 |
ENSDART00000185276
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr23_+_43177290 | 0.17 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr8_+_41038141 | 0.17 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr13_-_39159810 | 0.17 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr16_-_11766265 | 0.17 |
ENSDART00000141547
|
zgc:158701
|
zgc:158701 |
| chr13_+_12671513 | 0.17 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr7_-_41512999 | 0.17 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr3_-_49110710 | 0.17 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr4_+_5341592 | 0.17 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr22_-_3182965 | 0.17 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr18_-_15269272 | 0.17 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr16_+_11779534 | 0.17 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr7_+_3357973 | 0.17 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr10_-_22497980 | 0.16 |
ENSDART00000185549
|
CT573337.2
|
|
| chr24_+_39186940 | 0.16 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr22_+_9126557 | 0.16 |
ENSDART00000190669
|
si:ch211-156p11.1
|
si:ch211-156p11.1 |
| chr2_-_32768951 | 0.16 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr4_+_68939074 | 0.16 |
ENSDART00000180232
|
si:dkey-264f17.2
|
si:dkey-264f17.2 |
| chr14_-_14659023 | 0.16 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr25_+_23280220 | 0.16 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr10_+_25726694 | 0.16 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr9_+_40546677 | 0.16 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr22_+_9429710 | 0.16 |
ENSDART00000184089
|
si:ch211-11p18.6
|
si:ch211-11p18.6 |
| chr4_+_18804317 | 0.16 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr10_-_36254445 | 0.16 |
ENSDART00000123571
|
or110-2
|
odorant receptor, family D, subfamily 110, member 2 |
| chr14_+_6423973 | 0.16 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr25_-_10543682 | 0.16 |
ENSDART00000154262
|
tesmin
|
testis expressed metallothionein like protein |
| chr4_-_18850799 | 0.16 |
ENSDART00000151844
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr20_+_19119800 | 0.15 |
ENSDART00000151881
|
rp1l1b
|
retinitis pigmentosa 1-like 1b |
| chr9_-_42873700 | 0.15 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr24_+_37709191 | 0.15 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr23_+_25292147 | 0.15 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr14_+_20893065 | 0.15 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr4_+_56477160 | 0.15 |
ENSDART00000184152
|
si:ch211-237a4.2
|
si:ch211-237a4.2 |
| chr18_+_14277003 | 0.15 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr9_-_2936017 | 0.15 |
ENSDART00000102823
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr15_-_41503392 | 0.15 |
ENSDART00000169351
|
si:dkey-230i18.2
|
si:dkey-230i18.2 |
| chr14_-_12390724 | 0.15 |
ENSDART00000131343
|
magt1
|
magnesium transporter 1 |
| chr21_-_1644414 | 0.15 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr19_-_20163197 | 0.15 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr4_-_17741513 | 0.15 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr7_+_6879534 | 0.15 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr5_-_55914268 | 0.15 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr24_+_28561038 | 0.15 |
ENSDART00000147063
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr13_-_50200042 | 0.14 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr13_-_25196758 | 0.14 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr17_-_27266053 | 0.14 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr21_+_23108420 | 0.14 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr3_-_23513155 | 0.14 |
ENSDART00000170200
|
BX682558.1
|
|
| chr25_+_30105628 | 0.14 |
ENSDART00000187704
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr1_+_51721851 | 0.14 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr20_-_33959815 | 0.14 |
ENSDART00000133081
|
selp
|
selectin P |
| chr10_-_31015535 | 0.14 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr24_-_2381143 | 0.14 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr13_+_25412674 | 0.14 |
ENSDART00000113410
|
calhm1
|
calcium homeostasis modulator 1 |
| chr4_+_40383229 | 0.13 |
ENSDART00000126906
ENSDART00000191198 |
si:ch211-218h8.1
|
si:ch211-218h8.1 |
| chr24_-_26479841 | 0.13 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr14_+_30762131 | 0.13 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr18_-_24988645 | 0.13 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr2_+_21048661 | 0.13 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr15_-_41390410 | 0.13 |
ENSDART00000136164
|
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr10_+_18557102 | 0.13 |
ENSDART00000160157
|
CR388008.1
|
|
| chr2_+_6991208 | 0.13 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr22_+_8365905 | 0.13 |
ENSDART00000105945
|
si:dkey-19a16.11
|
si:dkey-19a16.11 |
| chr16_-_21888047 | 0.13 |
ENSDART00000162282
ENSDART00000124099 |
dicp3.3
|
diverse immunoglobulin domain-containing protein 3.3 |
| chr10_-_25823258 | 0.13 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr24_-_36727922 | 0.13 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr20_-_3238110 | 0.13 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr23_+_25291891 | 0.13 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr2_-_22569488 | 0.13 |
ENSDART00000158608
|
FQ377628.1
|
|
| chr12_-_44072716 | 0.12 |
ENSDART00000162509
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr21_+_21713531 | 0.12 |
ENSDART00000140501
|
or123-1
|
odorant receptor, family E, subfamily 123, member 1 |
| chr5_+_23242370 | 0.12 |
ENSDART00000051532
|
agtr2
|
angiotensin II receptor, type 2 |
| chr20_+_51833030 | 0.12 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr4_-_38519671 | 0.12 |
ENSDART00000183414
ENSDART00000187075 |
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr11_-_11336986 | 0.12 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr1_+_55239710 | 0.12 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr25_+_13620555 | 0.12 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr4_-_71551644 | 0.12 |
ENSDART00000168064
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr25_-_14637660 | 0.12 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr2_+_38731696 | 0.12 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr4_+_33871518 | 0.12 |
ENSDART00000185054
|
si:dkey-28i19.3
|
si:dkey-28i19.3 |
| chr21_-_3700334 | 0.12 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr8_+_52415603 | 0.12 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr3_-_33961589 | 0.12 |
ENSDART00000151533
|
ighd
|
immunoglobulin heavy constant delta |
| chr13_+_25455319 | 0.12 |
ENSDART00000145948
|
pkd2l1
|
polycystic kidney disease 2-like 1 |
| chr9_-_53666031 | 0.12 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr21_-_37194839 | 0.12 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr2_+_17569644 | 0.12 |
ENSDART00000135823
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr24_+_38208823 | 0.11 |
ENSDART00000187538
ENSDART00000190189 |
CU896602.3
|
|
| chr18_-_16792561 | 0.11 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr5_+_38619813 | 0.11 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr7_+_48555400 | 0.11 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr3_-_50139860 | 0.11 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr23_+_44049509 | 0.11 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr6_-_8399486 | 0.11 |
ENSDART00000189496
|
ptger1a
|
prostaglandin E receptor 1a (subtype EP1) |
| chr5_-_4204580 | 0.11 |
ENSDART00000049197
ENSDART00000132130 |
si:ch211-283g2.1
|
si:ch211-283g2.1 |
| chr12_-_4424424 | 0.11 |
ENSDART00000152314
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr1_+_15204633 | 0.11 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr25_+_11008419 | 0.11 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr21_-_43992027 | 0.11 |
ENSDART00000188612
|
cdx1b
|
caudal type homeobox 1 b |
| chr22_+_9091913 | 0.11 |
ENSDART00000186598
|
BX248395.1
|
|
| chr24_-_6671549 | 0.11 |
ENSDART00000114700
|
unm_sa821
|
un-named sa821 |
| chr9_-_5337515 | 0.11 |
ENSDART00000192701
|
tnfsf13b
|
TNF superfamily member 13b |
| chr6_+_54187643 | 0.11 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr15_+_37972671 | 0.11 |
ENSDART00000166764
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr25_+_37285737 | 0.11 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr22_+_22438783 | 0.11 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr22_+_9917007 | 0.10 |
ENSDART00000063318
|
blf
|
bloody fingers |
| chr10_+_8688678 | 0.10 |
ENSDART00000147568
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr12_-_4408828 | 0.10 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr1_-_2075529 | 0.10 |
ENSDART00000109983
|
oxgr1a.2
|
oxoglutarate (alpha-ketoglutarate) receptor 1a, tandem duplicate 2 |
| chr23_+_36052944 | 0.10 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr7_+_26545911 | 0.10 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr3_-_25118777 | 0.10 |
ENSDART00000188222
|
chadla
|
chondroadherin-like a |
| chr3_-_4303262 | 0.10 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr2_-_22575851 | 0.10 |
ENSDART00000182932
|
FQ377628.2
|
|
| chr7_+_4620586 | 0.10 |
ENSDART00000108797
|
zgc:162060
|
zgc:162060 |
| chr16_-_47465777 | 0.10 |
ENSDART00000190202
ENSDART00000181240 |
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr13_-_46882267 | 0.10 |
ENSDART00000161322
|
slc29a1a
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1a |
| chr18_+_45542981 | 0.10 |
ENSDART00000140357
|
kifc3
|
kinesin family member C3 |
| chr14_+_36454087 | 0.10 |
ENSDART00000180247
|
TENM3
|
si:dkey-237h12.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax10+pax6a+pax6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.3 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.2 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.2 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0030890 | B cell homeostasis(GO:0001782) positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) regulation of lipoprotein metabolic process(GO:0050746) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) regulation of protein lipidation(GO:1903059) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |