Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
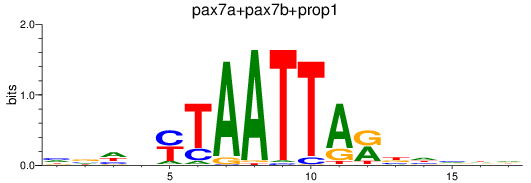
Results for pax7a+pax7b+prop1
Z-value: 0.45
Transcription factors associated with pax7a+pax7b+prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prop1
|
ENSDARG00000039756 | PROP paired-like homeobox 1 |
|
pax7b
|
ENSDARG00000070818 | paired box 7b |
|
pax7a
|
ENSDARG00000100398 | paired box 7a |
|
prop1
|
ENSDARG00000109950 | PROP paired-like homeobox 1 |
|
pax7a
|
ENSDARG00000111233 | paired box 7a |
|
prop1
|
ENSDARG00000114449 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax7a | dr11_v1_chr11_+_41242644_41242644 | 0.91 | 3.2e-02 | Click! |
| pax7b | dr11_v1_chr23_+_20931030_20931030 | 0.87 | 5.2e-02 | Click! |
| prop1 | dr11_v1_chr13_+_8677166_8677304 | 0.77 | 1.3e-01 | Click! |
Activity profile of pax7a+pax7b+prop1 motif
Sorted Z-values of pax7a+pax7b+prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9669717 | 0.61 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr2_+_50608099 | 0.41 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr12_+_18578597 | 0.41 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr17_-_12336987 | 0.34 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr13_-_29420885 | 0.33 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr20_-_29864390 | 0.33 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr16_+_47207691 | 0.33 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr4_+_4849789 | 0.32 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr4_-_815871 | 0.32 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr17_-_200316 | 0.31 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr24_-_31843173 | 0.31 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr21_-_20939488 | 0.29 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr6_+_44812444 | 0.29 |
ENSDART00000189478
ENSDART00000164653 ENSDART00000184534 ENSDART00000175473 |
chl1b
|
cell adhesion molecule L1-like b |
| chr16_-_12173554 | 0.29 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr16_-_12173399 | 0.29 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr17_+_23298928 | 0.29 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr19_-_9867001 | 0.28 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr7_-_58130703 | 0.28 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr25_-_19224298 | 0.28 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr15_-_33925851 | 0.28 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr21_+_28958471 | 0.28 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_+_47633438 | 0.28 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr21_+_31150438 | 0.27 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_+_44366556 | 0.27 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr10_+_45071603 | 0.27 |
ENSDART00000186505
ENSDART00000157573 |
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr9_-_35334642 | 0.27 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr21_+_31150773 | 0.25 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr17_-_36896560 | 0.25 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr10_+_45089820 | 0.25 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr13_+_15838151 | 0.25 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr2_-_21082695 | 0.25 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr9_-_43538328 | 0.24 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr3_-_6519691 | 0.24 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr4_-_22207873 | 0.24 |
ENSDART00000142140
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr10_+_15255198 | 0.23 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr22_-_26865361 | 0.23 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr11_+_30057762 | 0.23 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr4_+_9400012 | 0.23 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr18_-_48983690 | 0.22 |
ENSDART00000182359
|
FO681288.3
|
|
| chr12_+_48480632 | 0.22 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr2_-_37043905 | 0.22 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr13_-_11644806 | 0.21 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr2_-_42871286 | 0.21 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr16_+_2820340 | 0.21 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr23_+_8989168 | 0.21 |
ENSDART00000034380
|
xkr7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr2_+_29491314 | 0.21 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr7_+_23515966 | 0.21 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr20_+_19512727 | 0.21 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr1_+_6172786 | 0.21 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr23_-_24825863 | 0.20 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr23_+_35708730 | 0.20 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr3_-_21280373 | 0.20 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr10_+_42423318 | 0.20 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr14_-_49063157 | 0.20 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr25_-_19420949 | 0.20 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr8_+_18624658 | 0.19 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr18_+_26337869 | 0.19 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr9_-_47472998 | 0.19 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr1_+_14073891 | 0.19 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr24_-_7697274 | 0.18 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr6_+_39232245 | 0.18 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_+_5480327 | 0.18 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr3_-_23407720 | 0.18 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr21_-_39177564 | 0.18 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr3_-_35801146 | 0.18 |
ENSDART00000150911
|
caskin1
|
CASK interacting protein 1 |
| chr16_+_9726038 | 0.17 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr4_-_2219705 | 0.17 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr1_-_15797663 | 0.17 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr12_-_18393408 | 0.17 |
ENSDART00000159674
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr23_-_14990865 | 0.17 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr3_+_46459540 | 0.17 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr7_+_27898091 | 0.17 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr5_+_64842730 | 0.17 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr13_-_31452516 | 0.17 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr4_-_5019113 | 0.16 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr12_+_31735159 | 0.16 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr19_+_5604241 | 0.16 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr9_+_25776971 | 0.16 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr10_-_20706109 | 0.16 |
ENSDART00000124751
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr20_+_23173710 | 0.16 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr1_-_23157583 | 0.16 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr13_+_2442841 | 0.16 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr22_-_26865181 | 0.16 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr1_-_50859053 | 0.16 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr3_+_62000822 | 0.16 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr6_+_32882821 | 0.15 |
ENSDART00000075780
|
ghrhrb
|
growth hormone releasing hormone receptor b |
| chr10_+_21722892 | 0.15 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr23_-_9965148 | 0.15 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr22_+_10215558 | 0.15 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr10_+_25219728 | 0.15 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr15_-_18162647 | 0.15 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr21_+_27513859 | 0.15 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr8_+_31717175 | 0.15 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr9_+_54039006 | 0.15 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr8_+_17933475 | 0.15 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr15_-_30505607 | 0.15 |
ENSDART00000155212
|
msi2b
|
musashi RNA-binding protein 2b |
| chr3_-_23406964 | 0.15 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr15_-_22074315 | 0.14 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr25_-_13490744 | 0.14 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr19_-_5103141 | 0.14 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr16_+_40024883 | 0.14 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr16_-_22781446 | 0.14 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr5_-_5326010 | 0.14 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr20_+_18945057 | 0.14 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr20_-_47188966 | 0.14 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr18_+_38908903 | 0.14 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr19_+_19976990 | 0.14 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr16_-_28658341 | 0.14 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr3_-_34337969 | 0.14 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr9_-_14504834 | 0.14 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr12_+_16087077 | 0.14 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr25_+_5821910 | 0.13 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr21_-_42202792 | 0.13 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chrM_+_8130 | 0.13 |
ENSDART00000093609
|
mt-co2
|
cytochrome c oxidase II, mitochondrial |
| chr3_+_26019426 | 0.13 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_+_41540862 | 0.13 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr2_-_30668580 | 0.13 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_-_12407194 | 0.13 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr19_-_5103313 | 0.13 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr24_+_5912898 | 0.13 |
ENSDART00000132686
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr12_+_18681477 | 0.12 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr4_-_73756673 | 0.12 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr7_+_20629411 | 0.12 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr24_-_27400017 | 0.12 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr10_+_17235370 | 0.12 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr2_-_3403020 | 0.12 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr21_-_2209714 | 0.12 |
ENSDART00000163659
|
zgc:162971
|
zgc:162971 |
| chr15_+_23799461 | 0.12 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr18_+_24919614 | 0.12 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr10_-_28118035 | 0.12 |
ENSDART00000190836
ENSDART00000088852 |
med13a
|
mediator complex subunit 13a |
| chr16_-_34401412 | 0.12 |
ENSDART00000054020
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr13_+_6342410 | 0.12 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr14_+_45406299 | 0.12 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr12_-_41684729 | 0.12 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr8_-_50888806 | 0.12 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr24_+_5912635 | 0.12 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr9_+_12444494 | 0.12 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr10_-_26738209 | 0.12 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr19_-_42045372 | 0.11 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr10_+_42521943 | 0.11 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr13_-_39736938 | 0.11 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr5_-_12219572 | 0.11 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr2_+_42871831 | 0.11 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr5_+_1109098 | 0.11 |
ENSDART00000166268
|
LO017790.1
|
|
| chr24_-_5911973 | 0.11 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr2_+_57848844 | 0.11 |
ENSDART00000037279
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr18_-_15551360 | 0.11 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr6_+_35052721 | 0.11 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr11_-_27867024 | 0.11 |
ENSDART00000182136
ENSDART00000187587 |
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr9_+_38372216 | 0.11 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr6_+_21001264 | 0.10 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr4_+_25917915 | 0.10 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr25_-_6432463 | 0.10 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr10_-_5581487 | 0.10 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr17_+_5793248 | 0.10 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr24_-_6345965 | 0.10 |
ENSDART00000178287
|
zgc:174877
|
zgc:174877 |
| chr12_-_2522487 | 0.10 |
ENSDART00000022471
ENSDART00000145213 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr5_-_28625515 | 0.10 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr25_+_18889332 | 0.10 |
ENSDART00000017043
|
cax2
|
cation/H+ exchanger protein 2 |
| chr3_-_13461056 | 0.10 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr9_-_32177117 | 0.10 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr11_+_29770966 | 0.10 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr6_+_7421898 | 0.09 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr17_-_22021213 | 0.09 |
ENSDART00000078843
|
zmp:0000001102
|
zmp:0000001102 |
| chr2_-_17044959 | 0.09 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr15_-_45110011 | 0.09 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr17_-_40956035 | 0.09 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr6_+_40951227 | 0.09 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_-_42016693 | 0.09 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr15_-_16177603 | 0.09 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr15_-_17025212 | 0.09 |
ENSDART00000014465
|
hip1
|
huntingtin interacting protein 1 |
| chr2_+_11923615 | 0.09 |
ENSDART00000126118
|
trove2
|
TROVE domain family, member 2 |
| chr6_-_40029423 | 0.09 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr10_-_14556978 | 0.09 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr5_-_41494831 | 0.09 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr20_+_54274431 | 0.09 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr4_-_69189894 | 0.09 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr24_-_24724233 | 0.09 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr13_-_9841806 | 0.09 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr1_-_25370281 | 0.09 |
ENSDART00000050445
|
trim2a
|
tripartite motif containing 2a |
| chr8_-_13184989 | 0.09 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr1_+_44919221 | 0.09 |
ENSDART00000140402
|
wu:fc21g02
|
wu:fc21g02 |
| chr8_-_7567815 | 0.09 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr17_+_28533102 | 0.08 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr8_+_23521974 | 0.08 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr15_+_31820536 | 0.08 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr7_+_24645186 | 0.08 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
| chr16_-_21140097 | 0.08 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr8_-_53926228 | 0.08 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr8_-_30979494 | 0.08 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr1_+_52560549 | 0.08 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr16_-_41646164 | 0.08 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr7_+_24023653 | 0.08 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr16_-_13281380 | 0.08 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr4_+_16885854 | 0.08 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr12_-_33357655 | 0.08 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_-_3613558 | 0.08 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr16_+_16933002 | 0.08 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr23_+_43770149 | 0.08 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr24_+_26345609 | 0.08 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax7a+pax7b+prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0045938 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.3 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.0 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |