Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
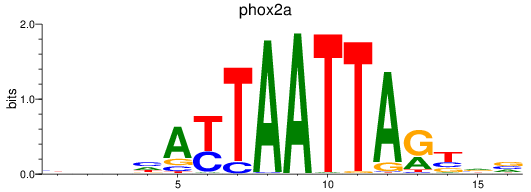
Results for phox2a
Z-value: 0.56
Transcription factors associated with phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2a
|
ENSDARG00000007406 | paired like homeobox 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2a | dr11_v1_chr15_+_47440477_47440477 | 0.79 | 1.2e-01 | Click! |
Activity profile of phox2a motif
Sorted Z-values of phox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_21129752 | 0.75 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr8_+_16025554 | 0.73 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_50608099 | 0.60 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr10_-_22918214 | 0.58 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr20_+_41756996 | 0.55 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr9_-_43538328 | 0.52 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr20_-_29864390 | 0.48 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr21_-_42100471 | 0.41 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr11_-_42554290 | 0.39 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr17_+_15433518 | 0.34 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_-_200316 | 0.34 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr17_+_15433671 | 0.33 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr4_-_1801519 | 0.32 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr16_-_25568512 | 0.31 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr5_+_4054704 | 0.28 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr3_-_20040636 | 0.28 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr10_+_42423318 | 0.24 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr21_-_13123176 | 0.24 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr21_+_32820175 | 0.23 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr12_-_33357655 | 0.21 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_+_5480327 | 0.21 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr18_+_34362608 | 0.18 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr1_-_44704261 | 0.17 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr20_-_28642061 | 0.16 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr6_-_35779348 | 0.15 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr9_-_32177117 | 0.14 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr2_+_2223837 | 0.14 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr24_+_28953089 | 0.14 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr7_-_30174882 | 0.12 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr15_-_43284021 | 0.11 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr1_-_19502322 | 0.10 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr5_+_19933356 | 0.10 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr1_-_15797663 | 0.09 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr15_-_14552101 | 0.09 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr13_+_31211050 | 0.09 |
ENSDART00000111705
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr5_+_60590796 | 0.08 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr10_-_11261565 | 0.07 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr15_-_22074315 | 0.07 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_+_37903790 | 0.05 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr2_-_9818640 | 0.05 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr3_-_56541723 | 0.04 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr19_+_2631565 | 0.03 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr6_+_13117598 | 0.03 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr16_+_28994709 | 0.03 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr6_+_24420523 | 0.03 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr12_+_26877381 | 0.02 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr18_+_28106139 | 0.01 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.2 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |