Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
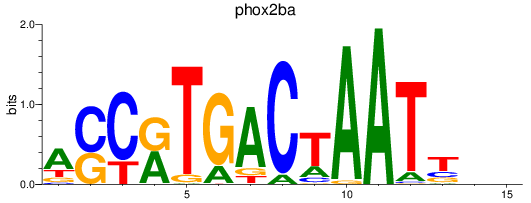
Results for phox2ba
Z-value: 0.43
Transcription factors associated with phox2ba
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2ba
|
ENSDARG00000075330 | paired like homeobox 2Ba |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2ba | dr11_v1_chr1_-_19079957_19079957 | 0.34 | 5.8e-01 | Click! |
Activity profile of phox2ba motif
Sorted Z-values of phox2ba motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77551860 | 0.54 |
ENSDART00000188176
|
AL935186.6
|
|
| chr2_-_53481912 | 0.49 |
ENSDART00000189610
|
hsd11b1lb
|
hydroxysteroid (11-beta) dehydrogenase 1-like b |
| chr24_-_27436319 | 0.43 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr9_-_54304684 | 0.43 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr4_-_4834347 | 0.28 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr3_+_6469754 | 0.25 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr14_+_45702521 | 0.24 |
ENSDART00000112470
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr4_-_4834617 | 0.23 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr15_+_5360407 | 0.21 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr13_-_12021566 | 0.19 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr1_+_58205904 | 0.19 |
ENSDART00000147324
|
si:dkey-222h21.12
|
si:dkey-222h21.12 |
| chr13_-_42560662 | 0.19 |
ENSDART00000124898
|
CR792417.1
|
|
| chr8_-_36370552 | 0.18 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr12_-_30043809 | 0.18 |
ENSDART00000150220
ENSDART00000075889 ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr17_+_45395846 | 0.18 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr13_-_34862452 | 0.17 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr1_+_57774454 | 0.17 |
ENSDART00000152507
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr24_-_26484298 | 0.17 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr14_-_45702721 | 0.17 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr18_+_27000850 | 0.16 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr1_+_26677406 | 0.16 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr10_+_26646104 | 0.16 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr18_+_27001115 | 0.16 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr2_+_44512324 | 0.15 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr10_+_26645953 | 0.15 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr1_+_33390286 | 0.14 |
ENSDART00000139004
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr19_+_4800068 | 0.14 |
ENSDART00000020686
|
st3gal1l
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like |
| chr22_-_15671525 | 0.14 |
ENSDART00000080286
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr1_+_26676758 | 0.13 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr9_-_21067971 | 0.13 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr11_+_13223625 | 0.13 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr2_-_51223658 | 0.12 |
ENSDART00000171770
|
pigrl4.2
|
polymeric immunoglobulin receptor-like 4.2 |
| chr7_+_8543317 | 0.12 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr18_+_27598755 | 0.12 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr1_-_33380011 | 0.11 |
ENSDART00000141347
ENSDART00000136383 |
cd99
|
CD99 molecule |
| chr2_+_55982940 | 0.10 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr5_-_44286987 | 0.10 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr19_-_32600138 | 0.10 |
ENSDART00000052101
|
zgc:91944
|
zgc:91944 |
| chr4_+_76509294 | 0.10 |
ENSDART00000099899
|
ms4a17a.17
|
membrane-spanning 4-domains, subfamily A, member 17A.17 |
| chr16_+_7662609 | 0.10 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr6_-_16493516 | 0.09 |
ENSDART00000022499
|
psmb3
|
proteasome subunit beta 3 |
| chr14_+_32839535 | 0.09 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr8_-_38355268 | 0.09 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr6_+_11850359 | 0.08 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr22_+_2533514 | 0.08 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr21_+_27189490 | 0.08 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr20_-_38801981 | 0.08 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr24_+_19542323 | 0.08 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr2_-_38282079 | 0.08 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr16_+_1471 | 0.08 |
ENSDART00000181972
|
CABZ01090785.1
|
|
| chr8_+_7854130 | 0.08 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr15_-_35960250 | 0.08 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr23_+_30048849 | 0.07 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr17_+_30587333 | 0.07 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr11_-_23459779 | 0.07 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr14_+_18781082 | 0.07 |
ENSDART00000108793
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr1_+_10074343 | 0.07 |
ENSDART00000190194
|
si:ch73-132f6.5
|
si:ch73-132f6.5 |
| chr6_+_16493755 | 0.07 |
ENSDART00000126318
|
si:ch73-193c12.2
|
si:ch73-193c12.2 |
| chr13_+_47623916 | 0.06 |
ENSDART00000109266
|
mertka
|
c-mer proto-oncogene tyrosine kinase a |
| chr13_+_42858228 | 0.06 |
ENSDART00000179763
|
cdh23
|
cadherin-related 23 |
| chr2_-_26590628 | 0.06 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr5_-_44286778 | 0.06 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr1_+_10074601 | 0.05 |
ENSDART00000168473
|
si:ch73-132f6.5
|
si:ch73-132f6.5 |
| chr19_-_32600638 | 0.05 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr17_+_24821627 | 0.05 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr5_-_38161033 | 0.05 |
ENSDART00000145907
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr21_+_18910502 | 0.04 |
ENSDART00000147567
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr23_-_14830627 | 0.04 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr19_-_10373630 | 0.04 |
ENSDART00000131517
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr17_+_21919131 | 0.03 |
ENSDART00000048136
|
htra1a
|
HtrA serine peptidase 1a |
| chr5_-_44829719 | 0.03 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr7_-_20796935 | 0.03 |
ENSDART00000162357
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr8_+_10862353 | 0.03 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr14_+_31493306 | 0.02 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr6_-_11800427 | 0.02 |
ENSDART00000126243
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr21_+_40239192 | 0.02 |
ENSDART00000135857
|
or115-9
|
odorant receptor, family F, subfamily 115, member 9 |
| chr2_-_9971609 | 0.01 |
ENSDART00000137924
ENSDART00000048655 ENSDART00000131613 |
zgc:55943
|
zgc:55943 |
| chr23_+_37090889 | 0.01 |
ENSDART00000074406
|
ubxn10
|
UBX domain protein 10 |
| chr6_-_9630081 | 0.01 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr15_+_36310442 | 0.01 |
ENSDART00000154678
|
gmnc
|
geminin coiled-coil domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2ba
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.0 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.4 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |