Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
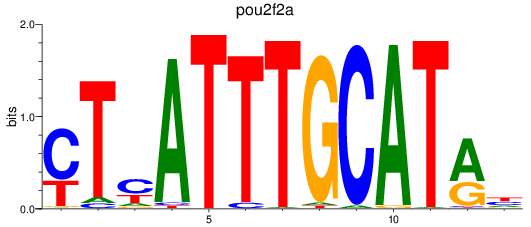
Results for pou2f2a
Z-value: 1.07
Transcription factors associated with pou2f2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f2a
|
ENSDARG00000019658 | POU class 2 homeobox 2a |
|
pou2f2a
|
ENSDARG00000036816 | POU class 2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f2a | dr11_v1_chr16_+_10918252_10918252 | -0.52 | 3.7e-01 | Click! |
Activity profile of pou2f2a motif
Sorted Z-values of pou2f2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_2669405 | 1.75 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr5_-_63302944 | 0.90 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr19_-_340641 | 0.87 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr17_+_26965351 | 0.85 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr19_-_40198478 | 0.82 |
ENSDART00000191736
|
grn2
|
granulin 2 |
| chr1_-_59169815 | 0.78 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr16_-_24550029 | 0.75 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr16_+_17714664 | 0.75 |
ENSDART00000149042
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr17_-_25382367 | 0.72 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr22_+_980290 | 0.66 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr17_-_2039511 | 0.66 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr16_+_17715243 | 0.65 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr8_+_20393250 | 0.65 |
ENSDART00000015038
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr23_+_7692042 | 0.62 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr5_+_58550291 | 0.61 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr23_+_23232136 | 0.61 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_+_26212621 | 0.58 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr14_-_15171435 | 0.58 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr5_+_15202495 | 0.56 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr4_-_4612116 | 0.56 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr5_-_42272517 | 0.55 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr7_-_57637779 | 0.52 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr19_+_19988869 | 0.51 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr23_-_31372639 | 0.51 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr14_-_38929885 | 0.50 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr7_-_21924372 | 0.50 |
ENSDART00000113652
|
si:dkey-85k7.10
|
si:dkey-85k7.10 |
| chr5_+_15203421 | 0.50 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr24_-_31904924 | 0.49 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr7_+_15736230 | 0.48 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr1_-_55118745 | 0.47 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr14_-_8080416 | 0.47 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr24_-_23758003 | 0.47 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr14_+_36889893 | 0.47 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr9_-_34396264 | 0.46 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr14_+_35428152 | 0.46 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr7_+_66565572 | 0.46 |
ENSDART00000180250
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr25_+_22320738 | 0.45 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr21_-_17290941 | 0.45 |
ENSDART00000147993
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr14_-_46198373 | 0.43 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr3_-_27646070 | 0.42 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr3_+_49021079 | 0.42 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr18_-_42830563 | 0.42 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr24_-_32452355 | 0.42 |
ENSDART00000146511
|
ccdc129
|
coiled-coil domain containing 129 |
| chr18_+_30567945 | 0.42 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr6_+_56147812 | 0.41 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_+_2230701 | 0.41 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr23_-_1017605 | 0.41 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr7_-_8315179 | 0.39 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr3_-_32337653 | 0.39 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr5_-_34993242 | 0.39 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr11_-_11575070 | 0.39 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr23_-_1017428 | 0.39 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr14_-_4145594 | 0.39 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr5_-_33287691 | 0.38 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr18_-_3520358 | 0.38 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr19_+_7864767 | 0.38 |
ENSDART00000137540
ENSDART00000151642 |
si:dkeyp-85e10.3
|
si:dkeyp-85e10.3 |
| chr9_+_33423170 | 0.38 |
ENSDART00000141542
|
si:dkey-216e24.9
|
si:dkey-216e24.9 |
| chr3_-_3209432 | 0.38 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr16_+_20895904 | 0.37 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr4_-_9780931 | 0.37 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr17_-_24916889 | 0.37 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr14_-_40821411 | 0.36 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr3_+_29476085 | 0.35 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr9_-_56272465 | 0.34 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_-_6553460 | 0.34 |
ENSDART00000172903
|
si:ch1073-220m6.1
|
si:ch1073-220m6.1 |
| chr21_-_34926619 | 0.34 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr7_+_4795707 | 0.33 |
ENSDART00000142704
|
si:dkey-28d5.4
|
si:dkey-28d5.4 |
| chr17_-_26867725 | 0.33 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr7_-_3649414 | 0.33 |
ENSDART00000064279
|
si:ch211-282j17.2
|
si:ch211-282j17.2 |
| chr21_+_31253048 | 0.32 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr5_+_54400971 | 0.32 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr24_+_39990695 | 0.31 |
ENSDART00000040281
|
BX323854.1
|
|
| chr5_+_26204561 | 0.30 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr13_-_25305179 | 0.30 |
ENSDART00000110064
|
plaua
|
plasminogen activator, urokinase a |
| chr9_+_54178475 | 0.30 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr23_-_32092443 | 0.30 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr12_-_4176329 | 0.29 |
ENSDART00000092746
|
si:dkey-32n7.7
|
si:dkey-32n7.7 |
| chr1_-_51720633 | 0.29 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr25_-_36248053 | 0.29 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T cells 3b |
| chr5_-_23749348 | 0.29 |
ENSDART00000140766
|
gbgt1l2
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 2 |
| chr20_-_25626693 | 0.29 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr3_-_34060153 | 0.29 |
ENSDART00000151610
|
ighv1-1
|
immunoglobulin heavy variable 1-1 |
| chr2_-_6482240 | 0.29 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr19_+_19775757 | 0.28 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr12_-_2940070 | 0.28 |
ENSDART00000112182
|
fam20a
|
family with sequence similarity 20, member A |
| chr8_+_7315625 | 0.28 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr17_-_26868169 | 0.28 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr4_+_11464255 | 0.28 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr7_+_4931366 | 0.28 |
ENSDART00000172384
ENSDART00000138168 ENSDART00000114543 |
si:dkey-83f18.3
|
si:dkey-83f18.3 |
| chr1_+_26667872 | 0.28 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr17_+_6563307 | 0.27 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr7_+_4987530 | 0.27 |
ENSDART00000132983
ENSDART00000137067 |
si:dkey-81n2.1
|
si:dkey-81n2.1 |
| chr7_+_3442834 | 0.27 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr20_+_25626479 | 0.27 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr10_+_10351685 | 0.27 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr7_-_32020858 | 0.27 |
ENSDART00000023568
|
kif18a
|
kinesin family member 18A |
| chr7_+_36041509 | 0.27 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr18_+_13248956 | 0.26 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr6_+_28428329 | 0.26 |
ENSDART00000188056
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr17_-_27382826 | 0.26 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr7_+_4922104 | 0.26 |
ENSDART00000135154
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr13_-_40411908 | 0.26 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr7_+_4624926 | 0.25 |
ENSDART00000137387
|
si:dkey-83f18.6
|
si:dkey-83f18.6 |
| chr10_-_1276046 | 0.25 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr13_-_30996072 | 0.25 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr14_+_46020571 | 0.25 |
ENSDART00000157617
ENSDART00000083928 |
c1qtnf2
|
C1q and TNF related 2 |
| chr5_+_9259971 | 0.25 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr7_+_5007310 | 0.25 |
ENSDART00000146083
|
si:ch211-272h9.3
|
si:ch211-272h9.3 |
| chr16_-_25608453 | 0.25 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr16_+_19637384 | 0.25 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr15_-_4580763 | 0.24 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr16_+_52999778 | 0.24 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr3_+_37112693 | 0.24 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr3_-_34113838 | 0.24 |
ENSDART00000151216
|
ighv4-2
|
immunoglobulin heavy variable 4-2 |
| chr14_-_32965169 | 0.24 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr16_-_2870522 | 0.24 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr2_+_10280645 | 0.24 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr6_+_43450221 | 0.24 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr14_+_35383267 | 0.24 |
ENSDART00000143818
|
clint1a
|
clathrin interactor 1a |
| chr11_+_18612421 | 0.24 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr1_-_25177086 | 0.23 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr25_+_5288665 | 0.23 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr1_-_55166511 | 0.23 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr1_-_56208151 | 0.22 |
ENSDART00000183325
|
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr18_-_7481036 | 0.22 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr16_-_43065164 | 0.22 |
ENSDART00000149431
|
sema4aa
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa |
| chr6_+_19950107 | 0.22 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr1_-_51038885 | 0.22 |
ENSDART00000035150
|
spast
|
spastin |
| chr7_+_44593756 | 0.22 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr3_+_28581397 | 0.22 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr7_-_3792765 | 0.21 |
ENSDART00000060333
|
si:dkey-28d5.12
|
si:dkey-28d5.12 |
| chr9_+_21820101 | 0.21 |
ENSDART00000145431
|
txndc9
|
thioredoxin domain containing 9 |
| chr7_-_3826639 | 0.21 |
ENSDART00000060330
|
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr12_+_37401331 | 0.21 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr8_-_19279784 | 0.21 |
ENSDART00000185892
|
zgc:77486
|
zgc:77486 |
| chr7_-_3792972 | 0.21 |
ENSDART00000146304
|
si:dkey-28d5.12
|
si:dkey-28d5.12 |
| chr8_+_20994433 | 0.20 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr25_-_16057539 | 0.20 |
ENSDART00000090388
|
cyb5r2
|
cytochrome b5 reductase 2 |
| chr12_+_4900981 | 0.20 |
ENSDART00000171507
|
cd79b
|
CD79b molecule, immunoglobulin-associated beta |
| chr3_+_41558682 | 0.20 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr3_-_34054081 | 0.20 |
ENSDART00000151590
|
ighv1-2
|
immunoglobulin heavy variable 1-2 |
| chr7_-_3723144 | 0.20 |
ENSDART00000138945
|
si:ch211-282j17.8
|
si:ch211-282j17.8 |
| chr21_-_11646878 | 0.20 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr7_+_4682458 | 0.20 |
ENSDART00000139271
|
si:ch211-225k7.2
|
si:ch211-225k7.2 |
| chr8_+_30747940 | 0.19 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr1_+_58137020 | 0.19 |
ENSDART00000180887
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr20_+_18994059 | 0.19 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr15_-_25556074 | 0.19 |
ENSDART00000124677
|
mmp20a
|
matrix metallopeptidase 20a (enamelysin) |
| chr8_+_28629741 | 0.19 |
ENSDART00000163915
|
si:dkey-109a10.2
|
si:dkey-109a10.2 |
| chr4_+_9592486 | 0.19 |
ENSDART00000080829
|
hspa14
|
heat shock protein 14 |
| chr9_-_40073255 | 0.19 |
ENSDART00000189139
|
IKZF2
|
si:zfos-1425h8.1 |
| chr25_+_8407892 | 0.19 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr7_-_3630019 | 0.18 |
ENSDART00000138164
ENSDART00000064277 |
si:dkey-192d15.3
|
si:dkey-192d15.3 |
| chr7_+_4911590 | 0.18 |
ENSDART00000038628
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr6_+_6779114 | 0.18 |
ENSDART00000163493
ENSDART00000143359 |
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr7_-_3850090 | 0.18 |
ENSDART00000113403
ENSDART00000137683 ENSDART00000049926 |
si:dkey-28d5.12
si:dkey-88n24.7
|
si:dkey-28d5.12 si:dkey-88n24.7 |
| chr13_+_45582391 | 0.18 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr2_-_51259371 | 0.18 |
ENSDART00000158049
ENSDART00000164916 ENSDART00000157792 |
si:ch211-215e19.6
|
si:ch211-215e19.6 |
| chr13_-_2112450 | 0.18 |
ENSDART00000189343
|
fam83b
|
family with sequence similarity 83, member B |
| chr8_+_50190742 | 0.18 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr7_+_4219234 | 0.18 |
ENSDART00000187891
ENSDART00000173246 ENSDART00000172808 |
si:ch211-63p21.4
|
si:ch211-63p21.4 |
| chr8_+_36570508 | 0.18 |
ENSDART00000145868
|
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr7_-_3884424 | 0.17 |
ENSDART00000145641
|
si:dkey-88n24.3
|
si:dkey-88n24.3 |
| chr7_-_3836896 | 0.17 |
ENSDART00000136227
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr10_+_4987766 | 0.17 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr11_+_18612166 | 0.17 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr7_-_29533127 | 0.17 |
ENSDART00000057289
|
anxa2b
|
annexin A2b |
| chr18_+_30998472 | 0.17 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr19_-_41518922 | 0.17 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr10_+_8690936 | 0.17 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr15_-_16012963 | 0.16 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr20_-_25542183 | 0.16 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr23_-_20126257 | 0.16 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr20_+_18993452 | 0.16 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr23_+_11347313 | 0.16 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr8_+_36570791 | 0.16 |
ENSDART00000145566
ENSDART00000180527 |
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr25_-_18739924 | 0.16 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr7_+_4865367 | 0.16 |
ENSDART00000139120
|
si:dkey-28d5.8
|
si:dkey-28d5.8 |
| chr23_+_12455295 | 0.16 |
ENSDART00000143752
ENSDART00000135785 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr10_+_42169982 | 0.16 |
ENSDART00000190905
|
CU467905.1
|
|
| chr1_-_45320126 | 0.16 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr7_-_3849926 | 0.16 |
ENSDART00000143064
|
si:dkey-88n24.7
|
si:dkey-88n24.7 |
| chr7_+_4854690 | 0.15 |
ENSDART00000131645
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr7_+_4694762 | 0.15 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr7_+_4592676 | 0.15 |
ENSDART00000141073
|
si:dkey-83f18.15
|
si:dkey-83f18.15 |
| chr24_-_8730913 | 0.15 |
ENSDART00000187228
ENSDART00000082349 ENSDART00000186660 |
tfap2a
|
transcription factor AP-2 alpha |
| chr7_-_3762896 | 0.15 |
ENSDART00000136711
ENSDART00000137140 |
si:ch211-282j17.11
|
si:ch211-282j17.11 |
| chr19_+_14573998 | 0.15 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr7_-_3836635 | 0.15 |
ENSDART00000104350
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr2_+_7851493 | 0.15 |
ENSDART00000176910
|
si:ch211-38m6.6
|
si:ch211-38m6.6 |
| chr9_-_9980704 | 0.15 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr17_+_15297398 | 0.14 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr7_+_4572727 | 0.14 |
ENSDART00000137830
|
si:dkey-83f18.7
|
si:dkey-83f18.7 |
| chr5_-_57686487 | 0.14 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr5_+_25585869 | 0.14 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr10_+_16756546 | 0.14 |
ENSDART00000168624
|
CABZ01024659.1
|
|
| chr6_+_52212927 | 0.14 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr3_-_34032019 | 0.14 |
ENSDART00000151779
|
ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr2_-_51303937 | 0.14 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr13_+_24396666 | 0.14 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 1.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 0.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.7 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0007440 | pancreatic D cell differentiation(GO:0003311) foregut morphogenesis(GO:0007440) |
| 0.0 | 0.4 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.6 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.2 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0048566 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.5 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0060061 | developmental induction(GO:0031128) Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) atrioventricular canal development(GO:0036302) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.2 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.4 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |