Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
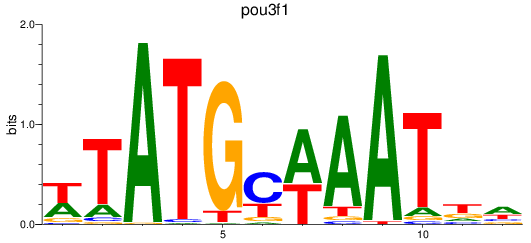
Results for pou3f1
Z-value: 0.97
Transcription factors associated with pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f1
|
ENSDARG00000009823 | POU class 3 homeobox 1 |
|
pou3f1
|
ENSDARG00000112302 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f1 | dr11_v1_chr16_+_33593116_33593116 | 0.95 | 1.2e-02 | Click! |
Activity profile of pou3f1 motif
Sorted Z-values of pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_30563115 | 1.39 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr22_+_20195280 | 1.29 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr23_+_28731379 | 0.99 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr18_-_36135799 | 0.97 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr23_+_44307996 | 0.97 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr19_-_28367413 | 0.90 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr19_-_30562648 | 0.90 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr3_-_5964557 | 0.86 |
ENSDART00000184738
|
BX284638.1
|
|
| chr21_-_27185915 | 0.86 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr9_-_18877597 | 0.85 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr23_+_36771593 | 0.85 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr16_-_12173399 | 0.84 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr7_+_40228422 | 0.83 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr6_-_13187168 | 0.81 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr16_-_12173554 | 0.80 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr12_+_48480632 | 0.76 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr16_-_43971258 | 0.75 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr7_-_38633867 | 0.73 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr22_+_5106751 | 0.73 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr21_+_41743493 | 0.72 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr10_+_22003750 | 0.72 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr2_-_34555945 | 0.72 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr3_-_6519691 | 0.71 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr17_-_15528597 | 0.69 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr3_+_33345348 | 0.69 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr16_+_45746549 | 0.66 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr23_+_28582865 | 0.63 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr13_+_33117528 | 0.63 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr6_-_13188667 | 0.62 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr5_-_24000211 | 0.62 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr5_-_16996482 | 0.60 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr6_+_4539953 | 0.60 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr22_-_38516922 | 0.58 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr2_+_24203229 | 0.57 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr16_+_43152727 | 0.56 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr4_-_8903240 | 0.55 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr8_+_28593707 | 0.55 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr18_-_12957451 | 0.54 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr22_-_7400471 | 0.54 |
ENSDART00000161215
|
si:dkey-57c15.1
|
si:dkey-57c15.1 |
| chr16_+_39159752 | 0.53 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr14_-_40389699 | 0.53 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr20_+_27298783 | 0.51 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr6_-_13308813 | 0.51 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr22_-_11954861 | 0.50 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
| chr12_+_46960579 | 0.49 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr2_+_2563192 | 0.48 |
ENSDART00000055713
|
CABZ01033000.1
|
|
| chr20_-_39188476 | 0.47 |
ENSDART00000152858
|
rcan2
|
regulator of calcineurin 2 |
| chr10_+_33895315 | 0.46 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr22_+_14051894 | 0.46 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr4_+_31259 | 0.45 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr5_+_30624183 | 0.45 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr1_+_46194333 | 0.45 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr7_-_24699985 | 0.45 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr22_+_413349 | 0.44 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr8_-_15398760 | 0.44 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr20_+_18225329 | 0.44 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr17_+_15882533 | 0.43 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr6_+_41096058 | 0.42 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr25_-_25736958 | 0.41 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr13_+_28705143 | 0.40 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr5_-_46896541 | 0.40 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr19_-_31707892 | 0.40 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr18_+_7286788 | 0.39 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr17_+_33719415 | 0.38 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr18_+_38749547 | 0.38 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr6_+_45692026 | 0.37 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr1_+_7679328 | 0.37 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr5_-_25406807 | 0.36 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr9_-_31915423 | 0.35 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr19_+_2726819 | 0.34 |
ENSDART00000187122
ENSDART00000112414 |
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr22_+_18786797 | 0.34 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr22_+_18389271 | 0.34 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr15_+_39096736 | 0.33 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr21_-_30254185 | 0.33 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr21_-_25573064 | 0.33 |
ENSDART00000134310
|
CR388166.1
|
|
| chr1_+_1599979 | 0.33 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr7_+_61050329 | 0.31 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr9_-_18735256 | 0.31 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr17_-_37052622 | 0.31 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr13_-_33822550 | 0.31 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr19_-_19556778 | 0.30 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr20_-_45060241 | 0.30 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr4_+_20255160 | 0.30 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr25_-_26673570 | 0.29 |
ENSDART00000154917
|
ciartb
|
circadian associated repressor of transcription b |
| chr24_-_40009446 | 0.29 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr17_+_20500229 | 0.29 |
ENSDART00000088490
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr15_+_42933236 | 0.28 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr16_-_26435431 | 0.28 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr11_-_35575791 | 0.28 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr15_-_30815826 | 0.27 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr1_+_31725154 | 0.27 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr21_+_5888641 | 0.27 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr9_-_21507083 | 0.27 |
ENSDART00000137922
|
aff3
|
AF4/FMR2 family, member 3 |
| chr13_+_29773153 | 0.27 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr6_-_19381993 | 0.27 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr18_+_660578 | 0.27 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr13_+_42406883 | 0.27 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr4_-_9579299 | 0.26 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr9_+_34331368 | 0.26 |
ENSDART00000147913
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr7_+_10701938 | 0.26 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr25_-_15045338 | 0.26 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr24_+_41931585 | 0.25 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr14_+_23874062 | 0.24 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr13_-_24877577 | 0.24 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr19_+_41464870 | 0.24 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr5_-_26323137 | 0.24 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr15_-_18361475 | 0.24 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr7_+_31074636 | 0.24 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr9_-_25444091 | 0.24 |
ENSDART00000002428
|
acvr2aa
|
activin A receptor type 2Aa |
| chr4_+_12292274 | 0.24 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr13_+_28618086 | 0.23 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr1_+_8984068 | 0.23 |
ENSDART00000122568
|
tlr2
|
toll-like receptor 2 |
| chr14_-_34700633 | 0.23 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_+_13824150 | 0.23 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr22_-_10397600 | 0.23 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr13_+_29778610 | 0.23 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr23_+_2825940 | 0.23 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr11_+_25634041 | 0.22 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr5_+_9147497 | 0.22 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr10_-_5821584 | 0.22 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr16_+_25074029 | 0.22 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr7_+_25920792 | 0.21 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr8_-_43574935 | 0.21 |
ENSDART00000051139
|
ncor2
|
nuclear receptor corepressor 2 |
| chr9_-_30502010 | 0.21 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr1_-_20593778 | 0.21 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr11_-_41130239 | 0.20 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr3_+_26223376 | 0.20 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr20_+_20751425 | 0.20 |
ENSDART00000177048
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr7_-_26497947 | 0.20 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr12_+_16087077 | 0.20 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr21_-_32467099 | 0.19 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr17_-_25831569 | 0.19 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr6_-_54111928 | 0.19 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr1_-_24918686 | 0.19 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr18_+_22606259 | 0.19 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr14_-_48939560 | 0.19 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr2_-_39036604 | 0.18 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr10_+_33744098 | 0.18 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr9_+_12444494 | 0.18 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr10_+_44940693 | 0.18 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr22_-_32392252 | 0.18 |
ENSDART00000148681
ENSDART00000137945 |
pcbp4
|
poly(rC) binding protein 4 |
| chr23_+_20518504 | 0.17 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr7_+_20965880 | 0.17 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr17_-_30473374 | 0.17 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr1_+_54013457 | 0.17 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr1_-_31350148 | 0.17 |
ENSDART00000085370
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr19_-_32042105 | 0.17 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr12_+_28986308 | 0.17 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr25_+_25737386 | 0.17 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr19_-_26770083 | 0.17 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr7_-_46777876 | 0.17 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr7_-_4036184 | 0.16 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr17_-_4245311 | 0.16 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr19_-_26769867 | 0.16 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr2_-_56131312 | 0.16 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr23_-_36934944 | 0.16 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr10_-_27196093 | 0.16 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr4_-_16406737 | 0.15 |
ENSDART00000013085
|
dcn
|
decorin |
| chr1_-_17587552 | 0.15 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr25_+_27923846 | 0.15 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr8_+_8238727 | 0.14 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr11_+_21586335 | 0.14 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr16_-_21620947 | 0.14 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_+_37545639 | 0.14 |
ENSDART00000173192
|
pcdh1b
|
protocadherin 1b |
| chr5_+_63785339 | 0.14 |
ENSDART00000050871
|
rgs3b
|
regulator of G protein signaling 3b |
| chr10_+_42521943 | 0.14 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr3_+_30190419 | 0.14 |
ENSDART00000157320
|
akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr3_-_21118969 | 0.13 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr6_+_18418651 | 0.13 |
ENSDART00000171097
ENSDART00000167798 |
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr20_-_2361226 | 0.13 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr24_-_37484123 | 0.13 |
ENSDART00000111623
|
cluap1
|
clusterin associated protein 1 |
| chr2_-_7185460 | 0.13 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr1_+_27825980 | 0.13 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr15_-_1622468 | 0.12 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr19_-_6239248 | 0.12 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr6_+_23935045 | 0.12 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr14_-_28001986 | 0.12 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr1_+_32051581 | 0.12 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr2_-_37684641 | 0.11 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr2_-_3616363 | 0.11 |
ENSDART00000144699
|
pter
|
phosphotriesterase related |
| chr7_+_69470442 | 0.11 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr8_+_31777633 | 0.11 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr17_+_26352372 | 0.11 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr23_-_893547 | 0.11 |
ENSDART00000136805
|
rbm10
|
RNA binding motif protein 10 |
| chr16_+_49089123 | 0.11 |
ENSDART00000187494
|
plcl2
|
phospholipase C like 2 |
| chr25_+_5604512 | 0.11 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr4_+_12013043 | 0.10 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr17_-_4245902 | 0.10 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr21_+_26390549 | 0.10 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr10_+_39304422 | 0.10 |
ENSDART00000019267
|
kcnj1b
|
potassium inwardly-rectifying channel, subfamily J, member 1b |
| chr10_-_26225548 | 0.10 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr5_+_69733096 | 0.10 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr16_+_34046733 | 0.10 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| chr21_-_35853245 | 0.10 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr22_-_18491813 | 0.10 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr1_-_40411944 | 0.10 |
ENSDART00000144135
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr23_-_26725637 | 0.10 |
ENSDART00000157844
|
hspg2
|
heparan sulfate proteoglycan 2 |
| chr21_+_8427059 | 0.10 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr7_+_60396957 | 0.09 |
ENSDART00000121545
|
brms1
|
breast cancer metastasis suppressor 1 |
| chr2_-_24317240 | 0.09 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.0 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.4 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.4 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.9 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 0.7 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.5 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.2 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.3 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.4 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.6 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.2 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |