Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
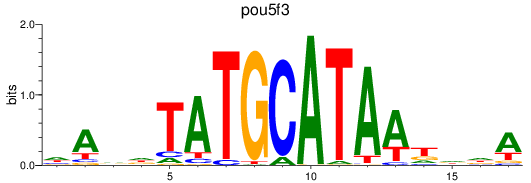
Results for pou5f3
Z-value: 0.69
Transcription factors associated with pou5f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou5f3
|
ENSDARG00000044774 | POU domain, class 5, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou5f3 | dr11_v1_chr21_-_13690712_13690712 | -0.24 | 7.0e-01 | Click! |
Activity profile of pou5f3 motif
Sorted Z-values of pou5f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_40994259 | 0.63 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr4_-_8903240 | 0.60 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr4_-_5302162 | 0.53 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr7_+_61050329 | 0.50 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr22_+_15507218 | 0.43 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr21_+_39361040 | 0.43 |
ENSDART00000085453
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr22_+_5106751 | 0.42 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr13_-_39830999 | 0.41 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr7_-_72155022 | 0.41 |
ENSDART00000187984
|
zmp:0000001168
|
zmp:0000001168 |
| chr11_-_36001495 | 0.40 |
ENSDART00000190330
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr8_-_22698651 | 0.38 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr8_+_7740004 | 0.37 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr19_-_43552252 | 0.37 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr8_-_16592491 | 0.32 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr24_-_21989406 | 0.32 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr20_-_34868814 | 0.32 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr11_+_7528599 | 0.30 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr24_-_20641000 | 0.28 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr25_+_26923193 | 0.26 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr9_+_12444494 | 0.24 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr8_+_43016714 | 0.22 |
ENSDART00000142671
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr11_+_19060278 | 0.21 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr24_+_3328354 | 0.17 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr13_+_18545819 | 0.15 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr2_-_20120904 | 0.15 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr11_+_19060079 | 0.14 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr13_+_31172833 | 0.13 |
ENSDART00000176378
|
CR931802.3
|
|
| chr7_-_24644893 | 0.13 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr16_-_31351419 | 0.13 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr24_-_25004553 | 0.12 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr24_+_17007407 | 0.12 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr8_-_44238526 | 0.12 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr7_+_33424044 | 0.11 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr18_+_7543347 | 0.08 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr4_+_6869847 | 0.06 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr24_-_7321928 | 0.04 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr11_+_13193747 | 0.04 |
ENSDART00000173336
|
si:dkey-9a6.4
|
si:dkey-9a6.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou5f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |