Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
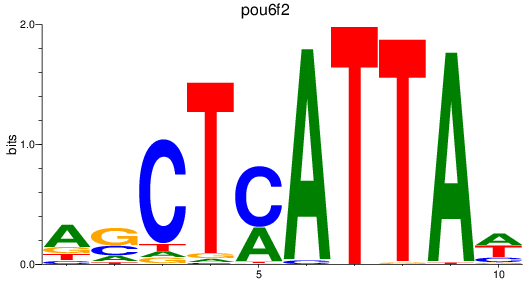
Results for pou6f2
Z-value: 1.17
Transcription factors associated with pou6f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f2
|
ENSDARG00000086362 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f2 | dr11_v1_chr24_+_10039165_10039165 | 0.80 | 1.0e-01 | Click! |
Activity profile of pou6f2 motif
Sorted Z-values of pou6f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_1185772 | 2.07 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr15_-_16098531 | 1.48 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr16_+_30301539 | 1.11 |
ENSDART00000186018
|
LO017848.1
|
|
| chr11_+_39672874 | 1.05 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr23_+_28731379 | 1.03 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr7_+_38717624 | 1.00 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr12_-_10275320 | 1.00 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr19_+_30662529 | 0.98 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr22_-_354592 | 0.95 |
ENSDART00000155769
|
tmem240b
|
transmembrane protein 240b |
| chr4_+_22365061 | 0.94 |
ENSDART00000039277
|
lhfpl3
|
LHFPL tetraspan subfamily member 3 |
| chr21_-_26918901 | 0.91 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr4_+_12615836 | 0.91 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr6_+_27146671 | 0.90 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr3_-_46817499 | 0.87 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_-_35787801 | 0.87 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr7_-_39203799 | 0.85 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr16_+_29303971 | 0.83 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr19_-_26869103 | 0.81 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr16_+_46111849 | 0.80 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr9_+_42063906 | 0.80 |
ENSDART00000048893
|
pcbp3
|
poly(rC) binding protein 3 |
| chr22_+_9472814 | 0.78 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr12_+_32729470 | 0.74 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr14_-_31465905 | 0.73 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr3_-_21288202 | 0.73 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr8_+_29267093 | 0.73 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr9_+_38888025 | 0.72 |
ENSDART00000148306
|
map2
|
microtubule-associated protein 2 |
| chr19_-_32641725 | 0.72 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr23_-_28141419 | 0.69 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr13_+_24552254 | 0.66 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr10_-_34870667 | 0.65 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr6_+_3004972 | 0.64 |
ENSDART00000186750
ENSDART00000183862 ENSDART00000191485 ENSDART00000171014 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr16_+_50741154 | 0.62 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr24_+_32411753 | 0.62 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr16_-_12173399 | 0.61 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr9_-_43538328 | 0.61 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr10_+_3507861 | 0.61 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr3_+_33341640 | 0.60 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr8_+_21588067 | 0.59 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr16_-_12173554 | 0.59 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr16_+_43152727 | 0.59 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr5_-_23362602 | 0.59 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr5_+_44064764 | 0.58 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr7_-_18416741 | 0.57 |
ENSDART00000097882
|
sstr1b
|
somatostatin receptor 1b |
| chr2_+_12302527 | 0.57 |
ENSDART00000091298
ENSDART00000182569 |
arhgap21b
|
Rho GTPase activating protein 21b |
| chr18_+_7283283 | 0.55 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr17_+_51499789 | 0.55 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr12_+_35654749 | 0.54 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr5_-_50641129 | 0.53 |
ENSDART00000190090
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr2_-_50372467 | 0.53 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr19_-_31402429 | 0.51 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr3_-_28250722 | 0.51 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr24_+_9744012 | 0.50 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr13_-_37545487 | 0.50 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr4_-_17629444 | 0.50 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr1_+_40023640 | 0.49 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr1_+_25801648 | 0.48 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_-_19364813 | 0.48 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr21_-_27185915 | 0.48 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr4_-_16124417 | 0.48 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr9_-_32753535 | 0.48 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr3_-_43356082 | 0.48 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr3_-_40054615 | 0.46 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr15_-_33172246 | 0.46 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr15_-_13208607 | 0.45 |
ENSDART00000184151
|
zgc:172282
|
zgc:172282 |
| chr13_-_31435137 | 0.44 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr7_-_27685365 | 0.43 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr8_+_51050554 | 0.42 |
ENSDART00000166249
|
DISP3
|
si:dkey-32e23.6 |
| chr5_-_35953472 | 0.40 |
ENSDART00000143448
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr4_+_12612145 | 0.40 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr25_+_6186823 | 0.40 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr2_+_29249204 | 0.39 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr25_+_13498188 | 0.39 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr11_+_24620742 | 0.38 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr21_-_39177564 | 0.38 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr11_+_38280454 | 0.37 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr11_-_43226255 | 0.37 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_-_27686021 | 0.37 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr4_-_1801519 | 0.36 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr18_-_19350792 | 0.35 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr7_+_31145386 | 0.35 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_+_17982737 | 0.34 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr20_+_40457599 | 0.34 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr9_-_6624663 | 0.34 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr13_+_42406883 | 0.34 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_+_33634186 | 0.34 |
ENSDART00000075626
|
htr1fb
|
5-hydroxytryptamine (serotonin) receptor 1Fb |
| chr11_-_37425407 | 0.33 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr15_+_22722684 | 0.33 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr20_+_4060839 | 0.32 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr11_+_19433936 | 0.32 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr18_-_2433011 | 0.32 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr24_+_29449690 | 0.31 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr10_-_26738209 | 0.31 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr18_+_34362608 | 0.30 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr6_+_37754763 | 0.30 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr17_+_11675362 | 0.30 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr17_-_37052622 | 0.29 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr24_+_40473032 | 0.29 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr9_-_14504834 | 0.27 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr6_+_49021703 | 0.27 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr4_+_12612723 | 0.27 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr15_-_14469704 | 0.26 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr11_+_7580079 | 0.25 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr23_-_28423048 | 0.24 |
ENSDART00000140448
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr21_-_37790727 | 0.24 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr19_-_5103313 | 0.24 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_+_10698549 | 0.23 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr23_-_6660985 | 0.22 |
ENSDART00000162405
|
CR450824.2
|
|
| chr5_-_48268049 | 0.22 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr12_+_41697664 | 0.22 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr24_-_21404367 | 0.21 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr19_+_8612839 | 0.21 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr4_+_3980247 | 0.21 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr6_-_10740365 | 0.21 |
ENSDART00000150942
|
sp3b
|
Sp3b transcription factor |
| chr3_+_57038033 | 0.20 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr25_-_21716326 | 0.20 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr18_+_14529005 | 0.19 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr11_+_33628104 | 0.19 |
ENSDART00000165318
|
thsd7bb
|
thrombospondin, type I, domain containing 7Bb |
| chr10_+_40284003 | 0.18 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr2_+_29249561 | 0.18 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr5_-_41933912 | 0.18 |
ENSDART00000097574
|
ncor1
|
nuclear receptor corepressor 1 |
| chr13_-_44630111 | 0.18 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr7_+_27976448 | 0.18 |
ENSDART00000181026
|
tub
|
tubby bipartite transcription factor |
| chr24_-_21923930 | 0.18 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr10_-_27199135 | 0.18 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr18_-_29962234 | 0.17 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr2_-_17044959 | 0.17 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr8_-_14080534 | 0.17 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr9_+_38502524 | 0.17 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr1_+_34203817 | 0.16 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr22_-_14696667 | 0.16 |
ENSDART00000180379
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr9_+_11034314 | 0.16 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr12_+_30367079 | 0.16 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr7_+_53498152 | 0.16 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr3_-_56541723 | 0.16 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr4_+_9467049 | 0.16 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr18_-_8398972 | 0.15 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr2_+_38892631 | 0.14 |
ENSDART00000134040
|
si:ch211-119o8.4
|
si:ch211-119o8.4 |
| chr19_+_9459050 | 0.14 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr2_-_21819421 | 0.14 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_23717165 | 0.14 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_37015185 | 0.14 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr7_-_31759602 | 0.14 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr13_+_25434932 | 0.13 |
ENSDART00000128562
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr17_-_45150763 | 0.13 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr7_+_36898850 | 0.13 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr15_+_24704798 | 0.13 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr3_-_55525627 | 0.13 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr15_+_7057050 | 0.13 |
ENSDART00000061828
|
foxl2a
|
forkhead box L2a |
| chr6_-_40713183 | 0.12 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr9_+_45789887 | 0.12 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr8_+_37755099 | 0.12 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr11_-_27702778 | 0.12 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr3_+_17537352 | 0.11 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr24_-_16979728 | 0.11 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr5_+_54497730 | 0.11 |
ENSDART00000157722
|
tmem203
|
transmembrane protein 203 |
| chr18_+_43365890 | 0.10 |
ENSDART00000173113
|
si:ch211-129p13.1
|
si:ch211-129p13.1 |
| chr14_-_26425416 | 0.10 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr9_-_49493305 | 0.10 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr7_-_4315859 | 0.09 |
ENSDART00000172762
|
si:ch211-63p21.8
|
si:ch211-63p21.8 |
| chr23_+_6077503 | 0.09 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr14_+_11290828 | 0.09 |
ENSDART00000184078
|
rlim
|
ring finger protein, LIM domain interacting |
| chr25_-_20049449 | 0.08 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr15_-_17870090 | 0.08 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr12_+_30367371 | 0.08 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr24_-_6628359 | 0.08 |
ENSDART00000169731
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr22_-_21897203 | 0.08 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr17_-_49977966 | 0.08 |
ENSDART00000183735
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr12_+_9183626 | 0.08 |
ENSDART00000020192
|
tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr15_-_38154616 | 0.07 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr22_-_30881738 | 0.07 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr22_+_2830703 | 0.07 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr23_+_33718602 | 0.06 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr16_+_42481447 | 0.06 |
ENSDART00000037401
|
herpud2
|
HERPUD family member 2 |
| chr22_+_25814941 | 0.06 |
ENSDART00000161120
|
vasna
|
vasorin a |
| chr2_+_56012016 | 0.06 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr15_+_21276735 | 0.06 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr21_+_25231160 | 0.06 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr9_+_21793565 | 0.05 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr20_-_9436521 | 0.05 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr16_-_2414063 | 0.05 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr13_+_24662238 | 0.05 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr13_-_3370638 | 0.05 |
ENSDART00000029649
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr12_-_26406323 | 0.05 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr21_-_3853204 | 0.05 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_-_41638039 | 0.04 |
ENSDART00000144525
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr8_-_23776399 | 0.04 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr4_+_72723304 | 0.04 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr13_+_3938526 | 0.03 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr7_-_31759394 | 0.03 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr10_-_24391716 | 0.03 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr2_-_37312927 | 0.02 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr3_-_30434016 | 0.02 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr6_+_4033832 | 0.02 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr11_+_33284837 | 0.02 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr17_-_29902187 | 0.02 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr20_+_13781779 | 0.02 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr7_-_30174882 | 0.01 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr8_-_4327473 | 0.01 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr17_+_12865746 | 0.01 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr16_+_25107344 | 0.01 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.3 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.4 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 2.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 1.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.4 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.6 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.8 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.6 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.4 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 4.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.4 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |