Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
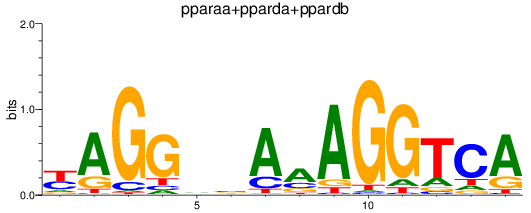
Results for pparaa+pparda+ppardb
Z-value: 1.19
Transcription factors associated with pparaa+pparda+ppardb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ppardb
|
ENSDARG00000009473 | peroxisome proliferator-activated receptor delta b |
|
pparaa
|
ENSDARG00000031777 | peroxisome proliferator-activated receptor alpha a |
|
pparda
|
ENSDARG00000044525 | peroxisome proliferator-activated receptor delta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ppardb | dr11_v1_chr8_+_23709280_23709280 | -0.52 | 3.7e-01 | Click! |
| pparda | dr11_v1_chr22_+_1006573_1006573 | -0.40 | 5.0e-01 | Click! |
| pparaa | dr11_v1_chr4_-_18635005_18635005 | 0.17 | 7.8e-01 | Click! |
Activity profile of pparaa+pparda+ppardb motif
Sorted Z-values of pparaa+pparda+ppardb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_11201096 | 1.47 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr9_-_43082945 | 0.94 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr6_-_39765546 | 0.93 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_39764995 | 0.85 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_-_19006631 | 0.85 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr1_-_17797802 | 0.83 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr11_-_6974022 | 0.80 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr7_+_34297271 | 0.78 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr15_-_12500938 | 0.78 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr20_+_47953047 | 0.78 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr3_+_27770110 | 0.74 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr7_+_34296789 | 0.72 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_30545895 | 0.70 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr14_-_29826659 | 0.68 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr13_-_37127970 | 0.59 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_+_28602423 | 0.59 |
ENSDART00000182350
|
crot
|
carnitine O-octanoyltransferase |
| chr23_+_17417539 | 0.56 |
ENSDART00000182605
|
BX649300.2
|
|
| chr21_+_15704556 | 0.53 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr20_-_47953524 | 0.52 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr19_+_40861853 | 0.49 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr10_+_35417099 | 0.46 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr23_-_24488696 | 0.46 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr17_-_32370047 | 0.46 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr6_+_60036767 | 0.45 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr14_-_48103207 | 0.45 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr15_+_404891 | 0.44 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr16_+_28601974 | 0.44 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr1_+_40802454 | 0.42 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr17_-_20711735 | 0.42 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr20_-_34164278 | 0.40 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr3_-_53486169 | 0.37 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr17_+_30546579 | 0.37 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr24_+_9590188 | 0.34 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr3_-_34561624 | 0.34 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr2_-_26476030 | 0.33 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr20_+_15015557 | 0.32 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr24_+_2961098 | 0.32 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr16_-_50229193 | 0.30 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr24_+_81527 | 0.29 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr8_-_2529878 | 0.28 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr13_+_9842276 | 0.25 |
ENSDART00000041700
ENSDART00000141406 |
prdx3
|
peroxiredoxin 3 |
| chr5_+_58421536 | 0.25 |
ENSDART00000173410
|
ccdc15
|
coiled-coil domain containing 15 |
| chr10_+_11265387 | 0.24 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr7_-_24046999 | 0.24 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr1_-_46859398 | 0.23 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr8_-_979735 | 0.22 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr20_-_38836161 | 0.22 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr9_+_30626416 | 0.21 |
ENSDART00000147813
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr20_-_48604199 | 0.21 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr15_-_6946286 | 0.20 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr14_+_989733 | 0.19 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr17_+_30450163 | 0.19 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr20_-_52967878 | 0.18 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr18_+_16744307 | 0.18 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr18_+_27926839 | 0.17 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr20_-_48604621 | 0.17 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr19_+_2631565 | 0.16 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr19_+_19652439 | 0.15 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr5_-_41875834 | 0.15 |
ENSDART00000133592
|
si:dkey-65b12.10
|
si:dkey-65b12.10 |
| chr5_+_43870389 | 0.14 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr8_+_2530065 | 0.14 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr11_+_24957858 | 0.14 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr15_+_31820536 | 0.13 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr1_-_35113974 | 0.13 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr13_+_28819768 | 0.13 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr8_-_20291922 | 0.13 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr8_-_8698607 | 0.12 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr3_+_30257582 | 0.11 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr12_-_684200 | 0.11 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr13_+_28821841 | 0.11 |
ENSDART00000179900
|
CU639469.1
|
|
| chr23_-_16737161 | 0.11 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr15_+_28268135 | 0.10 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr5_+_38276582 | 0.10 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr22_+_22888 | 0.10 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr7_+_23940933 | 0.10 |
ENSDART00000173628
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr19_-_7043355 | 0.10 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr3_+_58167288 | 0.08 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr17_+_6538733 | 0.08 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_+_17356482 | 0.08 |
ENSDART00000063629
|
nitr3r.1l
|
novel immune-type receptor 3, related 1-like |
| chr23_-_16734009 | 0.08 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr22_-_10774735 | 0.07 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr5_-_35137365 | 0.07 |
ENSDART00000141218
|
fcho2
|
FCH domain only 2 |
| chr19_+_32158010 | 0.06 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr22_+_18469004 | 0.06 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr5_-_41860556 | 0.05 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr6_-_30863046 | 0.05 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr12_-_5120175 | 0.04 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr20_-_25877793 | 0.04 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr20_+_46286082 | 0.04 |
ENSDART00000073519
|
taar14f
|
trace amine associated receptor 14f |
| chr10_-_41400049 | 0.04 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr22_+_9069081 | 0.04 |
ENSDART00000187842
|
BX248395.1
|
|
| chr1_-_52292235 | 0.04 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr22_-_38274188 | 0.04 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr12_+_26670778 | 0.03 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_+_48675347 | 0.03 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr25_+_36347126 | 0.03 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr10_+_36441124 | 0.02 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr20_+_1088658 | 0.02 |
ENSDART00000162991
|
BX537249.1
|
|
| chr21_+_26720803 | 0.02 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr12_-_44199316 | 0.02 |
ENSDART00000170378
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr5_-_30984010 | 0.02 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr18_-_3456117 | 0.02 |
ENSDART00000158508
|
myo7aa
|
myosin VIIAa |
| chr12_+_46708920 | 0.02 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr15_+_47618221 | 0.02 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr22_-_3595439 | 0.02 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr8_+_8699085 | 0.01 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr12_-_5120339 | 0.01 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr1_-_58868306 | 0.01 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr18_-_12957451 | 0.01 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr9_+_500052 | 0.01 |
ENSDART00000166707
|
CU984600.1
|
|
| chr23_+_44633858 | 0.00 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr17_-_12196865 | 0.00 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pparaa+pparda+ppardb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.8 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 0.3 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 1.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0071711 | basement membrane organization(GO:0071711) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.4 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 0.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.0 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.2 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.4 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |