Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
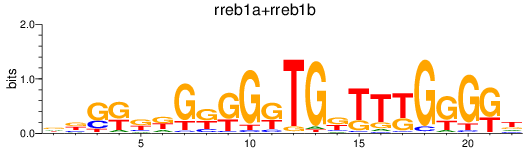
Results for rreb1a+rreb1b
Z-value: 1.10
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1a | dr11_v1_chr24_-_2381143_2381143 | 0.97 | 6.9e-03 | Click! |
| rreb1b | dr11_v1_chr2_+_21000334_21000355 | 0.72 | 1.7e-01 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_258883 | 0.97 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr23_+_44580254 | 0.74 |
ENSDART00000185069
|
pfn1
|
profilin 1 |
| chr4_+_77735212 | 0.73 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr19_-_5380770 | 0.71 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr11_+_36243774 | 0.53 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr7_+_31838320 | 0.51 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr19_-_5364649 | 0.51 |
ENSDART00000004812
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr19_-_5358443 | 0.50 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr9_+_30294096 | 0.50 |
ENSDART00000026551
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr13_-_45523026 | 0.45 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr18_+_5547185 | 0.43 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr8_+_554531 | 0.43 |
ENSDART00000193623
|
FO704758.2
|
|
| chr13_+_22675802 | 0.41 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr13_-_51846224 | 0.40 |
ENSDART00000184663
|
LT631684.2
|
|
| chr15_-_47479119 | 0.40 |
ENSDART00000164957
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr17_+_53311618 | 0.39 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr4_-_277081 | 0.38 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr17_+_53418445 | 0.38 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr1_+_227241 | 0.38 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr17_+_15388479 | 0.38 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr9_+_10692905 | 0.34 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr12_-_54375 | 0.34 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr14_-_9522364 | 0.33 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr8_-_554540 | 0.32 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr11_+_45233348 | 0.31 |
ENSDART00000173150
|
tmc6b
|
transmembrane channel-like 6b |
| chr4_+_90048 | 0.31 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr23_-_38054 | 0.31 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr18_+_46151505 | 0.31 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr7_-_73752955 | 0.31 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr3_-_1434135 | 0.30 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr21_+_28445052 | 0.30 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr22_+_661505 | 0.30 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr15_-_28618502 | 0.30 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr7_+_74150839 | 0.30 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr14_-_33585809 | 0.29 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr6_-_442163 | 0.29 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr1_+_59538755 | 0.29 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr2_-_51700709 | 0.28 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr1_-_59243542 | 0.28 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr4_+_76619791 | 0.27 |
ENSDART00000184042
|
ms4a17a.8
|
membrane-spanning 4-domains, subfamily A, member 17A.8 |
| chr12_+_2677303 | 0.27 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr23_-_912817 | 0.26 |
ENSDART00000192851
|
rhoac
|
ras homolog gene family, member Ac |
| chr18_-_49078428 | 0.26 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr20_-_1847433 | 0.25 |
ENSDART00000164211
|
CABZ01065425.1
|
|
| chr13_-_11536951 | 0.25 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr21_-_44009169 | 0.25 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr22_+_1186866 | 0.25 |
ENSDART00000167710
|
eps8l3a
|
EPS8-like 3a |
| chr5_-_72289648 | 0.25 |
ENSDART00000037691
|
tbx5a
|
T-box 5a |
| chr25_+_5044780 | 0.25 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr9_+_36946340 | 0.25 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr25_-_1124851 | 0.24 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr25_-_37422859 | 0.24 |
ENSDART00000156018
|
cd44b
|
CD44 molecule (Indian blood group) b |
| chr11_+_45347961 | 0.24 |
ENSDART00000172962
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr3_+_37196258 | 0.24 |
ENSDART00000187944
ENSDART00000185896 |
fmnl1a
|
formin-like 1a |
| chr7_+_35229645 | 0.24 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_55002583 | 0.24 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr6_+_392815 | 0.24 |
ENSDART00000163142
|
cyth4b
|
cytohesin 4b |
| chr22_+_10090673 | 0.23 |
ENSDART00000186680
|
si:dkey-102c8.3
|
si:dkey-102c8.3 |
| chr1_-_7930679 | 0.23 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr10_+_8527196 | 0.23 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr17_+_53311243 | 0.23 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr9_+_56449505 | 0.23 |
ENSDART00000187725
|
CABZ01079480.1
|
|
| chr7_-_55454406 | 0.23 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr19_-_13923091 | 0.22 |
ENSDART00000169993
ENSDART00000158992 |
stx12
|
syntaxin 12 |
| chr16_-_30885838 | 0.22 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr9_-_56420087 | 0.22 |
ENSDART00000112016
|
actr3
|
ARP3 actin related protein 3 homolog |
| chr6_-_21189295 | 0.22 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr4_+_77948517 | 0.22 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr10_-_43294933 | 0.22 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr4_+_76722754 | 0.21 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr1_-_48933 | 0.21 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr3_+_301479 | 0.21 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr16_-_54405976 | 0.21 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr1_-_9104631 | 0.21 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr3_-_30685401 | 0.20 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr2_+_45441563 | 0.20 |
ENSDART00000151882
|
si:ch211-66k16.27
|
si:ch211-66k16.27 |
| chr25_+_1732838 | 0.20 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr5_-_42180205 | 0.20 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr4_+_77948970 | 0.20 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr15_+_47362728 | 0.20 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr13_+_51853716 | 0.20 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr14_+_15620640 | 0.19 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr4_+_76748500 | 0.19 |
ENSDART00000075607
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr11_-_40147032 | 0.19 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr14_-_34059681 | 0.19 |
ENSDART00000003993
|
itk
|
IL2 inducible T cell kinase |
| chr13_+_41022502 | 0.19 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr3_-_60027255 | 0.19 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr10_+_22772084 | 0.18 |
ENSDART00000144845
ENSDART00000165222 |
tmem88a
|
transmembrane protein 88 a |
| chr17_-_122680 | 0.18 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr22_-_834106 | 0.18 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr14_+_21905492 | 0.18 |
ENSDART00000054437
|
tbc1d10c
|
TBC1 domain family, member 10C |
| chr9_+_22764235 | 0.18 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr8_-_1909840 | 0.18 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr18_-_226800 | 0.18 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr18_-_50845804 | 0.18 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr14_-_1432875 | 0.18 |
ENSDART00000164408
ENSDART00000159774 |
zgc:152774
|
zgc:152774 |
| chr24_-_41478917 | 0.18 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr1_+_59533317 | 0.17 |
ENSDART00000166821
|
sp6
|
Sp6 transcription factor |
| chr6_+_27338512 | 0.17 |
ENSDART00000155004
|
klhl30
|
kelch-like family member 30 |
| chr2_+_36007449 | 0.17 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr21_+_170038 | 0.17 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr20_+_27096430 | 0.17 |
ENSDART00000153358
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr22_-_5918670 | 0.17 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr8_+_3379815 | 0.17 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr16_+_7991274 | 0.17 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr7_+_35229805 | 0.16 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_+_176583 | 0.16 |
ENSDART00000168760
ENSDART00000160425 |
LAMP1
|
lysosomal associated membrane protein 1 |
| chr23_-_9768700 | 0.16 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr5_-_26181863 | 0.16 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr8_-_2602572 | 0.16 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr22_+_20169352 | 0.16 |
ENSDART00000169055
ENSDART00000061617 |
hmg20b
|
high mobility group 20B |
| chr16_+_53387085 | 0.16 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr14_+_26759332 | 0.16 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr9_-_23033818 | 0.15 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr21_+_6212844 | 0.15 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr23_+_22873415 | 0.15 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr16_+_68069 | 0.15 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr12_+_28117365 | 0.15 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr15_-_47468085 | 0.15 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr1_-_16688965 | 0.15 |
ENSDART00000079001
ENSDART00000178599 |
frg1
|
FSHD region gene 1 |
| chr19_+_917852 | 0.15 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr6_-_18601304 | 0.15 |
ENSDART00000170810
|
cd7al
|
cd7 antigen-like |
| chr4_-_18211 | 0.15 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr9_-_23807032 | 0.14 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr5_+_21891305 | 0.14 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr21_+_25625026 | 0.14 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr5_-_69621227 | 0.14 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr3_-_1400309 | 0.14 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr6_+_27514465 | 0.14 |
ENSDART00000128985
ENSDART00000079397 |
ryk
|
receptor-like tyrosine kinase |
| chr8_+_22355909 | 0.14 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr15_-_12500938 | 0.14 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr2_-_57264262 | 0.14 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr23_-_1348933 | 0.14 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr25_-_37489917 | 0.13 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr10_-_45058886 | 0.13 |
ENSDART00000159347
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr7_+_22293894 | 0.13 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr10_-_40826657 | 0.13 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr3_-_1190132 | 0.13 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr12_+_34273240 | 0.13 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr12_-_4540564 | 0.12 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr16_+_43347966 | 0.12 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr24_+_42131564 | 0.12 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr9_-_216527 | 0.12 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr19_+_7835025 | 0.12 |
ENSDART00000026276
|
cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr17_+_8925232 | 0.12 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr5_+_20589472 | 0.12 |
ENSDART00000130911
|
cmklr1
|
chemokine-like receptor 1 |
| chr9_+_23714406 | 0.12 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr12_-_1951233 | 0.12 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr17_+_15899039 | 0.12 |
ENSDART00000154181
|
syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr19_+_638339 | 0.12 |
ENSDART00000148705
|
tert
|
telomerase reverse transcriptase |
| chr5_+_337215 | 0.11 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
| chr8_+_46010838 | 0.11 |
ENSDART00000143126
|
si:ch211-119d14.2
|
si:ch211-119d14.2 |
| chr17_+_132555 | 0.11 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr20_+_48739154 | 0.11 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr19_+_7929704 | 0.11 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr6_+_36821621 | 0.11 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr2_-_32486080 | 0.11 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr18_+_173603 | 0.11 |
ENSDART00000185918
|
larp6a
|
La ribonucleoprotein domain family, member 6a |
| chr5_+_59116068 | 0.11 |
ENSDART00000154978
|
si:ch211-284f22.3
|
si:ch211-284f22.3 |
| chr7_+_50849142 | 0.11 |
ENSDART00000073806
|
pcolceb
|
procollagen C-endopeptidase enhancer b |
| chr23_-_46201008 | 0.11 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr19_+_2619444 | 0.10 |
ENSDART00000169483
|
fam126a
|
family with sequence similarity 126, member A |
| chr20_+_46513651 | 0.10 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr2_-_6373829 | 0.10 |
ENSDART00000081633
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr12_+_23892972 | 0.10 |
ENSDART00000152852
|
svila
|
supervillin a |
| chr9_+_56194410 | 0.10 |
ENSDART00000168530
|
LO018176.1
|
|
| chr22_+_30089054 | 0.10 |
ENSDART00000185369
|
add3a
|
adducin 3 (gamma) a |
| chr1_+_27690 | 0.10 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr19_+_42983613 | 0.10 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr25_+_5288665 | 0.10 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr20_-_35246150 | 0.10 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr3_+_23703704 | 0.10 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr17_-_5860222 | 0.10 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr12_+_48681601 | 0.10 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr2_-_51511494 | 0.10 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr2_-_8648440 | 0.10 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr7_-_5162292 | 0.10 |
ENSDART00000084218
|
zgc:195075
|
zgc:195075 |
| chr10_+_8481660 | 0.10 |
ENSDART00000109559
|
BX682234.1
|
|
| chr20_+_4392687 | 0.10 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr21_+_15601100 | 0.10 |
ENSDART00000180558
ENSDART00000145454 |
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr2_-_59285085 | 0.10 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr12_+_2676878 | 0.10 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr3_-_1408487 | 0.09 |
ENSDART00000100845
|
bglap
|
bone gamma-carboxyglutamate (gla) protein |
| chr4_-_1403077 | 0.09 |
ENSDART00000189813
|
CABZ01068469.1
|
|
| chr18_-_26715156 | 0.09 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr21_-_226071 | 0.09 |
ENSDART00000160667
|
nup54
|
nucleoporin 54 |
| chr19_+_48117995 | 0.09 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr24_-_32582378 | 0.09 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr13_-_36761379 | 0.09 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr14_+_34514336 | 0.09 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr6_-_43677125 | 0.09 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr1_+_14406098 | 0.09 |
ENSDART00000030083
|
chic2
|
cysteine-rich hydrophobic domain 2 |
| chr24_+_31334209 | 0.09 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr2_+_47754419 | 0.09 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr15_+_2534740 | 0.09 |
ENSDART00000138469
|
cux1b
|
cut-like homeobox 1b |
| chr5_-_30080332 | 0.09 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr7_+_73308566 | 0.09 |
ENSDART00000187039
ENSDART00000174244 |
CABZ01081777.1
|
|
| chr22_-_6144428 | 0.09 |
ENSDART00000106118
|
si:dkey-19a16.4
|
si:dkey-19a16.4 |
| chr19_+_7930004 | 0.09 |
ENSDART00000160410
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr15_+_714203 | 0.09 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.9 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.2 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.3 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.3 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.1 | 0.2 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:2000404 | regulation of T cell migration(GO:2000404) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |