Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
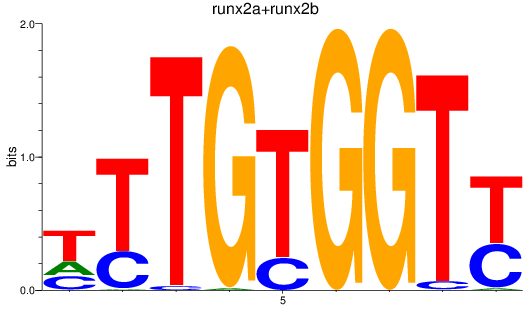
Results for runx2a+runx2b
Z-value: 1.28
Transcription factors associated with runx2a+runx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx2a
|
ENSDARG00000040261 | RUNX family transcription factor 2a |
|
runx2b
|
ENSDARG00000059233 | RUNX family transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx2b | dr11_v1_chr20_-_44055095_44055123 | 0.86 | 6.3e-02 | Click! |
| runx2a | dr11_v1_chr17_+_5351922_5351922 | 0.29 | 6.4e-01 | Click! |
Activity profile of runx2a+runx2b motif
Sorted Z-values of runx2a+runx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_23277939 | 1.43 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr19_+_30633453 | 1.30 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr24_-_4973765 | 1.15 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr5_-_23277385 | 0.94 |
ENSDART00000134982
|
plp1b
|
proteolipid protein 1b |
| chr7_+_25858380 | 0.90 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr2_-_44282796 | 0.89 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr2_-_44283554 | 0.89 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr7_+_46368520 | 0.84 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr13_+_27316632 | 0.84 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr23_-_15878879 | 0.84 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr13_+_27316934 | 0.83 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr19_-_9867001 | 0.77 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr15_+_29088426 | 0.71 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr15_+_39977461 | 0.70 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr1_+_15137901 | 0.69 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr25_+_34984333 | 0.65 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr15_+_19544052 | 0.62 |
ENSDART00000062560
|
zgc:77784
|
zgc:77784 |
| chr21_+_7582036 | 0.61 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr22_-_33679277 | 0.61 |
ENSDART00000169948
|
FO904977.1
|
|
| chr8_+_14778292 | 0.61 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr9_+_38883388 | 0.60 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr7_-_33829824 | 0.60 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr2_-_44280061 | 0.60 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr8_+_36803415 | 0.60 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr2_+_22694382 | 0.58 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr5_-_6377865 | 0.56 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr16_+_31853919 | 0.56 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr20_+_5564042 | 0.55 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr1_+_49266886 | 0.55 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr15_-_12545683 | 0.55 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr9_+_42066030 | 0.55 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr17_+_26815021 | 0.54 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr5_+_44190974 | 0.54 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr19_+_16222618 | 0.54 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr3_-_22212764 | 0.53 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr7_-_38633867 | 0.53 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr9_+_38888025 | 0.52 |
ENSDART00000148306
|
map2
|
microtubule-associated protein 2 |
| chr7_-_12065668 | 0.52 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr8_+_25173317 | 0.50 |
ENSDART00000142006
|
gpr61
|
G protein-coupled receptor 61 |
| chr13_+_7292061 | 0.50 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr16_+_50741154 | 0.49 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr1_+_41854298 | 0.48 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr14_-_46616487 | 0.47 |
ENSDART00000105417
ENSDART00000166550 ENSDART00000105418 |
prom1a
|
prominin 1a |
| chr22_+_20195280 | 0.47 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr7_+_23515966 | 0.47 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr4_+_41602 | 0.47 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr4_+_17336557 | 0.46 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr4_-_2525916 | 0.46 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr19_+_37857936 | 0.46 |
ENSDART00000189289
|
nxph1
|
neurexophilin 1 |
| chr2_-_31936966 | 0.45 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr6_+_13933464 | 0.45 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr3_+_54168007 | 0.45 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr23_-_36724575 | 0.44 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr4_+_20255160 | 0.44 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr20_+_38725662 | 0.43 |
ENSDART00000181873
|
uts1
|
urotensin 1 |
| chr10_-_17103651 | 0.43 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr6_+_38381957 | 0.43 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr4_+_11375894 | 0.43 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr10_+_21797276 | 0.43 |
ENSDART00000169105
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr7_+_38770167 | 0.42 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr5_-_28606916 | 0.42 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr3_+_28576173 | 0.42 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr10_+_19338255 | 0.42 |
ENSDART00000113737
|
lrrtm4l1
|
leucine rich repeat transmembrane neuronal 4 like 1 |
| chr21_-_21096437 | 0.42 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr13_-_23095006 | 0.42 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr3_+_15271943 | 0.42 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr21_-_42100471 | 0.42 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_8428736 | 0.42 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr13_+_35856463 | 0.41 |
ENSDART00000171056
ENSDART00000017202 |
kcnk1b
|
potassium channel, subfamily K, member 1b |
| chr18_+_18612388 | 0.41 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr15_-_45538773 | 0.41 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr17_-_46457622 | 0.41 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr17_+_13031497 | 0.41 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr13_-_51247529 | 0.40 |
ENSDART00000191774
ENSDART00000083788 |
AL929217.1
|
|
| chr2_-_50298337 | 0.40 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr8_-_17067364 | 0.40 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr4_-_8882572 | 0.40 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr24_-_31843173 | 0.40 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr11_+_24994705 | 0.39 |
ENSDART00000129211
|
zgc:92107
|
zgc:92107 |
| chr7_-_72605673 | 0.39 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr9_+_17984358 | 0.39 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr5_-_32445835 | 0.39 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr23_-_637347 | 0.39 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr19_-_3482138 | 0.38 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr2_-_22286828 | 0.38 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr11_-_44999858 | 0.38 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr9_+_7909490 | 0.38 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr21_+_21201346 | 0.38 |
ENSDART00000142961
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr3_+_15773991 | 0.37 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr3_-_28428198 | 0.37 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr3_-_36440705 | 0.37 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr3_+_14339728 | 0.36 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr13_+_11440389 | 0.36 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_+_29600917 | 0.36 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr16_+_11483811 | 0.36 |
ENSDART00000169012
ENSDART00000173042 |
grik5
|
glutamate receptor, ionotropic, kainate 5 |
| chr20_-_30035326 | 0.35 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr10_-_7974155 | 0.35 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr24_-_24848612 | 0.35 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr7_-_40993456 | 0.35 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr14_-_14746767 | 0.35 |
ENSDART00000183755
ENSDART00000190938 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr23_+_27789795 | 0.34 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr2_+_24722771 | 0.34 |
ENSDART00000147207
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr16_-_32672883 | 0.34 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr23_+_28582865 | 0.34 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr15_-_24869826 | 0.34 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr6_+_13806466 | 0.34 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr21_-_35324091 | 0.33 |
ENSDART00000185042
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr11_-_27057572 | 0.33 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr8_-_50888806 | 0.33 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr18_+_45573416 | 0.33 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr2_-_40135942 | 0.33 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr13_+_40019001 | 0.33 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr15_-_34213898 | 0.33 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr25_+_19954576 | 0.33 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr15_-_34214440 | 0.33 |
ENSDART00000167052
|
etv1
|
ets variant 1 |
| chr13_-_10261383 | 0.32 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr9_-_37749973 | 0.32 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr6_-_16394528 | 0.32 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr5_-_2752573 | 0.32 |
ENSDART00000168121
|
lzts3a
|
leucine zipper, putative tumor suppressor family member 3a |
| chr16_-_9869056 | 0.32 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr25_+_21324588 | 0.31 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr19_+_42693855 | 0.31 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr10_+_10738880 | 0.31 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr5_-_38342992 | 0.31 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr12_+_32159272 | 0.31 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr17_-_200316 | 0.31 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr1_+_10018466 | 0.31 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr3_+_37197686 | 0.30 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr19_+_5146460 | 0.30 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr7_+_72882446 | 0.30 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr12_+_18543783 | 0.30 |
ENSDART00000148326
ENSDART00000134530 |
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr4_+_3980247 | 0.30 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr5_-_52277643 | 0.30 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr9_-_28867562 | 0.30 |
ENSDART00000189597
ENSDART00000060321 |
zgc:91818
|
zgc:91818 |
| chr1_+_6817292 | 0.30 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr3_-_1204341 | 0.30 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr6_+_23810529 | 0.30 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr6_-_10305918 | 0.29 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr25_-_7925269 | 0.29 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr25_+_20119466 | 0.29 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr4_-_9533345 | 0.29 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr15_+_46108548 | 0.29 |
ENSDART00000099023
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr24_+_37080771 | 0.29 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr6_+_55357188 | 0.29 |
ENSDART00000158219
|
pltp
|
phospholipid transfer protein |
| chr16_-_21591533 | 0.29 |
ENSDART00000159870
|
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr18_-_21267103 | 0.29 |
ENSDART00000136285
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr20_-_31781941 | 0.29 |
ENSDART00000139417
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr6_+_29860776 | 0.28 |
ENSDART00000028406
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr5_+_61657702 | 0.28 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr23_+_26979174 | 0.28 |
ENSDART00000018654
|
rnd1b
|
Rho family GTPase 1b |
| chr8_+_8735285 | 0.28 |
ENSDART00000165655
|
elk1
|
ELK1, member of ETS oncogene family |
| chr25_-_32449235 | 0.28 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr25_-_7925019 | 0.28 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr2_+_31948352 | 0.28 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr13_-_29424454 | 0.28 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr6_-_12314475 | 0.28 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr7_+_31051603 | 0.27 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr14_-_2163454 | 0.27 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr3_-_37476278 | 0.27 |
ENSDART00000083442
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr1_-_55810730 | 0.27 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr15_+_6117502 | 0.27 |
ENSDART00000188293
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr7_+_63325819 | 0.27 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr10_+_26515946 | 0.27 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr18_+_45573251 | 0.26 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr5_+_62374092 | 0.26 |
ENSDART00000082965
|
CU928117.1
|
|
| chr9_-_46842179 | 0.26 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr3_+_36160979 | 0.26 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr7_-_18598661 | 0.26 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr8_+_13849999 | 0.26 |
ENSDART00000143784
|
doc2d
|
double C2-like domains, delta |
| chr15_+_17446796 | 0.26 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr25_+_31929325 | 0.26 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr11_+_37144328 | 0.26 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr15_+_37197494 | 0.26 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr15_-_35252522 | 0.25 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr14_+_24845941 | 0.25 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr23_-_7799184 | 0.25 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr9_+_35860975 | 0.25 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr1_+_2101541 | 0.25 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr8_-_39859688 | 0.25 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr19_-_5812319 | 0.25 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr17_+_691453 | 0.25 |
ENSDART00000159271
|
fancm
|
Fanconi anemia, complementation group M |
| chr2_-_42871286 | 0.25 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr16_-_22863348 | 0.25 |
ENSDART00000147609
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr4_-_15420452 | 0.25 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr21_+_34992550 | 0.25 |
ENSDART00000109041
ENSDART00000135400 |
tmprss15
|
transmembrane protease, serine 15 |
| chr23_-_27822920 | 0.25 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr15_+_42767803 | 0.25 |
ENSDART00000164879
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr18_-_40508528 | 0.24 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr10_+_21796477 | 0.24 |
ENSDART00000176255
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr8_+_26432677 | 0.24 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr24_-_18877118 | 0.24 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr5_+_23597144 | 0.24 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr21_+_33503835 | 0.24 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr23_-_29376859 | 0.24 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr13_+_17468161 | 0.23 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr8_+_29856013 | 0.23 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr24_-_10919588 | 0.23 |
ENSDART00000131204
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr12_-_48992527 | 0.23 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr20_+_18225329 | 0.23 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr18_-_41375120 | 0.23 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
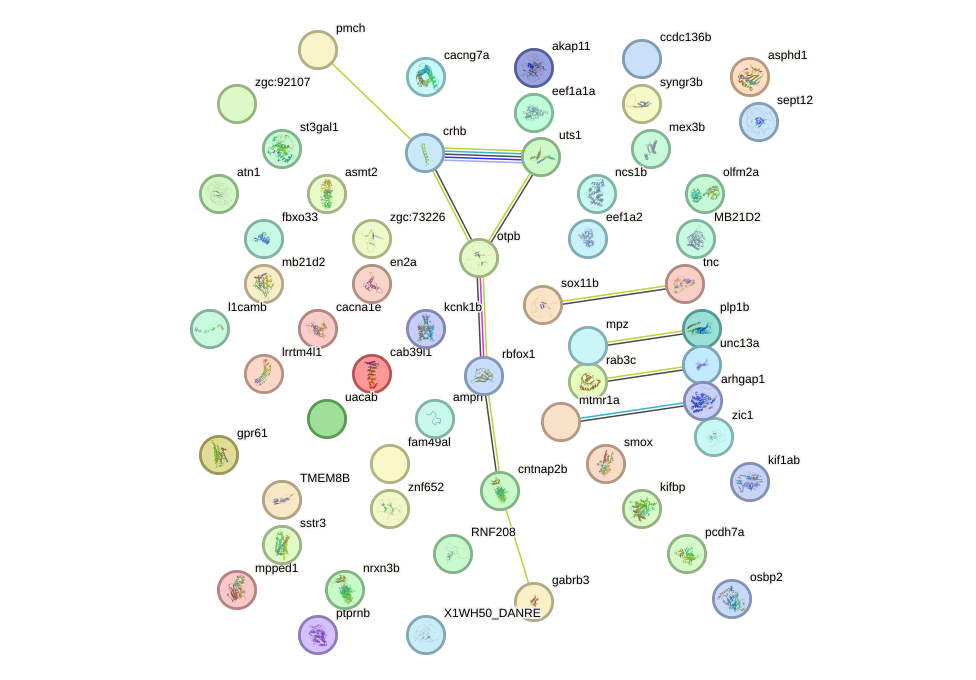
Network of associatons between targets according to the STRING database.
First level regulatory network of runx2a+runx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.6 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.5 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.2 | 1.2 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 2.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.6 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.8 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.3 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.6 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.1 | GO:0002837 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.2 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.6 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 1.1 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 2.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |