Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
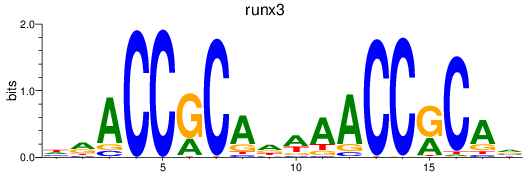
Results for runx3
Z-value: 0.95
Transcription factors associated with runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx3
|
ENSDARG00000052826 | RUNX family transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx3 | dr11_v1_chr13_-_45123051_45123051 | 0.85 | 6.9e-02 | Click! |
Activity profile of runx3 motif
Sorted Z-values of runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_43711918 | 1.58 |
ENSDART00000193110
|
CU914622.2
|
|
| chr19_-_5380770 | 0.79 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr12_-_43819253 | 0.68 |
ENSDART00000160261
ENSDART00000170045 ENSDART00000159106 |
si:ch73-329n5.6
|
si:ch73-329n5.6 |
| chr16_+_23431189 | 0.56 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr15_+_6117502 | 0.56 |
ENSDART00000188293
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr20_-_34670236 | 0.53 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr15_-_2657508 | 0.52 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr6_-_141564 | 0.51 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr16_+_28754403 | 0.49 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr4_-_13518381 | 0.47 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr15_+_37589698 | 0.43 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr19_+_37118547 | 0.41 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr5_-_71995108 | 0.38 |
ENSDART00000124587
|
fam78ab
|
family with sequence similarity 78, member Ab |
| chr20_-_48485354 | 0.32 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr23_-_36313431 | 0.31 |
ENSDART00000125860
|
nfe2
|
nuclear factor, erythroid 2 |
| chr12_+_46883785 | 0.31 |
ENSDART00000008312
|
fam53b
|
family with sequence similarity 53, member B |
| chr11_-_30636163 | 0.31 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr5_+_24059598 | 0.29 |
ENSDART00000051547
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr6_+_44163727 | 0.29 |
ENSDART00000064878
|
gxylt2
|
glucoside xylosyltransferase 2 |
| chr11_-_40519886 | 0.28 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr12_+_18744610 | 0.27 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr1_-_39859626 | 0.27 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr6_-_2154137 | 0.27 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr19_-_72398 | 0.27 |
ENSDART00000165587
|
dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr15_-_4037626 | 0.25 |
ENSDART00000160905
|
P2RY14
|
purinergic receptor P2Y14 |
| chr14_-_16476863 | 0.24 |
ENSDART00000089021
|
canx
|
calnexin |
| chr4_+_15983541 | 0.23 |
ENSDART00000043660
|
copg2
|
coatomer protein complex, subunit gamma 2 |
| chr20_-_14012859 | 0.23 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr22_-_30973791 | 0.22 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr8_+_18830759 | 0.22 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr19_+_4068134 | 0.22 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr15_-_37589600 | 0.22 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr7_+_25858380 | 0.21 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr19_+_2546775 | 0.21 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr7_+_52712807 | 0.20 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr19_+_4062101 | 0.19 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr25_-_8201983 | 0.19 |
ENSDART00000006579
|
saal1
|
serum amyloid A-like 1 |
| chr5_+_27429872 | 0.19 |
ENSDART00000087894
|
zgc:165555
|
zgc:165555 |
| chr16_-_54907588 | 0.18 |
ENSDART00000185709
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_+_28910762 | 0.18 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr23_+_36308717 | 0.17 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr1_-_43727418 | 0.16 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr19_+_2590182 | 0.16 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr18_+_808911 | 0.16 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr7_+_24888910 | 0.15 |
ENSDART00000173994
|
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr2_-_183992 | 0.15 |
ENSDART00000126704
ENSDART00000191283 ENSDART00000034783 |
zgc:113518
|
zgc:113518 |
| chr15_-_9321921 | 0.15 |
ENSDART00000159676
ENSDART00000033411 |
trappc4
|
trafficking protein particle complex 4 |
| chr22_+_29994093 | 0.15 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr8_-_17184482 | 0.14 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr22_+_11144153 | 0.14 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr10_-_309894 | 0.13 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr20_-_53321499 | 0.13 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr2_-_52841762 | 0.13 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr17_-_27382826 | 0.13 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr22_-_11520405 | 0.13 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr19_-_40906161 | 0.13 |
ENSDART00000004514
|
bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr8_-_18899427 | 0.13 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr2_+_4146606 | 0.13 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr24_-_18659147 | 0.12 |
ENSDART00000166039
|
zgc:66014
|
zgc:66014 |
| chr18_-_50524017 | 0.12 |
ENSDART00000150013
ENSDART00000149912 |
cd276
|
CD276 molecule |
| chr25_-_225964 | 0.11 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr13_+_8255106 | 0.11 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr17_+_37310663 | 0.10 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr20_-_2725930 | 0.10 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr2_+_16173999 | 0.10 |
ENSDART00000177639
ENSDART00000157601 ENSDART00000190186 ENSDART00000045933 |
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr15_+_29292154 | 0.10 |
ENSDART00000137817
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr22_-_14161309 | 0.10 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr12_-_3978306 | 0.10 |
ENSDART00000149473
ENSDART00000114857 |
ppp4cb
|
protein phosphatase 4, catalytic subunit b |
| chr16_-_7228276 | 0.09 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr6_+_39930825 | 0.09 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr16_+_35870456 | 0.08 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr25_-_210730 | 0.06 |
ENSDART00000187580
|
FP236318.1
|
|
| chr9_+_37141836 | 0.06 |
ENSDART00000024555
|
gli2a
|
GLI family zinc finger 2a |
| chr3_-_3372259 | 0.05 |
ENSDART00000140482
|
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr19_+_42219165 | 0.05 |
ENSDART00000163192
|
CU896644.1
|
|
| chr14_-_24332786 | 0.05 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr3_-_31086770 | 0.05 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr18_-_50523399 | 0.05 |
ENSDART00000033591
|
cd276
|
CD276 molecule |
| chr1_+_46598764 | 0.04 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr19_-_24443867 | 0.04 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr2_+_49864219 | 0.04 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr7_+_17938128 | 0.04 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr3_-_25369557 | 0.04 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr6_+_38896158 | 0.04 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr3_+_6291635 | 0.03 |
ENSDART00000185055
ENSDART00000157707 |
si:ch211-12p12.2
|
si:ch211-12p12.2 |
| chr1_-_18446161 | 0.03 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr5_+_62009765 | 0.03 |
ENSDART00000134335
|
akap10
|
A kinase (PRKA) anchor protein 10 |
| chr2_-_56348727 | 0.02 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr6_-_10740365 | 0.02 |
ENSDART00000150942
|
sp3b
|
Sp3b transcription factor |
| chr5_+_62009930 | 0.02 |
ENSDART00000082872
|
akap10
|
A kinase (PRKA) anchor protein 10 |
| chr13_+_45524475 | 0.01 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr24_-_14212521 | 0.01 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr1_+_39859782 | 0.01 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr15_+_24005289 | 0.01 |
ENSDART00000088648
|
myo18ab
|
myosin XVIIIAb |
| chr7_+_17937922 | 0.01 |
ENSDART00000147189
|
mta2
|
metastasis associated 1 family, member 2 |
| chr5_+_71995056 | 0.00 |
ENSDART00000180574
|
PLPP7 (1 of many)
|
phospholipid phosphatase 7 (inactive) |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |