Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
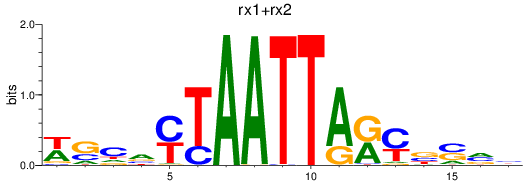
Results for rx1+rx2
Z-value: 0.85
Transcription factors associated with rx1+rx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx2
|
ENSDARG00000040321 | retinal homeobox gene 2 |
|
rx1
|
ENSDARG00000071684 | retinal homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx1 | dr11_v1_chr22_-_5006119_5006119 | -0.17 | 7.8e-01 | Click! |
| rx2 | dr11_v1_chr2_-_55853943_55853943 | -0.07 | 9.1e-01 | Click! |
Activity profile of rx1+rx2 motif
Sorted Z-values of rx1+rx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_6751405 | 0.93 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr21_+_6751760 | 0.72 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr10_-_34002185 | 0.60 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr15_-_5815006 | 0.53 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr18_-_5598958 | 0.53 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr3_-_26017831 | 0.53 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr20_+_43925266 | 0.52 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr16_+_23984179 | 0.48 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr7_+_6652967 | 0.45 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr18_-_40708537 | 0.41 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr5_+_37903790 | 0.38 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr2_+_6255434 | 0.33 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr16_-_31622777 | 0.26 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr3_+_17537352 | 0.24 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr5_+_32260502 | 0.24 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr16_-_31351419 | 0.24 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr17_+_24821627 | 0.23 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr22_+_7738966 | 0.21 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr16_-_13818061 | 0.21 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr17_+_24318753 | 0.19 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr12_+_47698356 | 0.17 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr7_+_54642005 | 0.17 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr15_-_14552101 | 0.16 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr25_+_11008419 | 0.15 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr24_-_25144441 | 0.15 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr23_+_12134839 | 0.14 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr4_-_9891874 | 0.14 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr3_-_23407720 | 0.13 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr22_+_19407531 | 0.13 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr7_+_34620418 | 0.13 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr14_+_901847 | 0.13 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr8_-_53044300 | 0.12 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr22_+_5123479 | 0.12 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr6_+_36839509 | 0.12 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr6_-_7720332 | 0.12 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr20_-_43663494 | 0.12 |
ENSDART00000144564
|
BX470188.1
|
|
| chr14_-_14659023 | 0.12 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr17_-_53329704 | 0.12 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr24_+_38301080 | 0.11 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr4_+_9467049 | 0.11 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr9_-_52962521 | 0.11 |
ENSDART00000170419
|
CU855885.1
|
|
| chr7_+_46368520 | 0.10 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr8_-_24502997 | 0.09 |
ENSDART00000184801
ENSDART00000179794 |
BX296549.1
|
|
| chr16_-_27677930 | 0.09 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr22_-_20011476 | 0.09 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr4_-_4387012 | 0.09 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr15_-_19051152 | 0.08 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr11_+_2855430 | 0.08 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr14_-_4170654 | 0.08 |
ENSDART00000188347
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr14_+_40874608 | 0.08 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr22_+_23359369 | 0.08 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr9_+_45493341 | 0.08 |
ENSDART00000145616
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr1_+_35985813 | 0.08 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr23_+_43255328 | 0.08 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr2_+_10007113 | 0.08 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr5_-_25733745 | 0.07 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr15_+_45640906 | 0.07 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr16_+_35379720 | 0.07 |
ENSDART00000170497
|
si:dkey-34d22.2
|
si:dkey-34d22.2 |
| chr9_-_11676491 | 0.07 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr8_+_28695914 | 0.07 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr5_+_58550795 | 0.07 |
ENSDART00000192282
|
pou2f3
|
POU class 2 homeobox 3 |
| chr20_-_13774826 | 0.07 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr7_-_33960170 | 0.07 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr6_+_40922572 | 0.07 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr21_-_30082414 | 0.06 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr21_-_2341937 | 0.06 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr12_-_41684729 | 0.06 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr4_-_61691066 | 0.06 |
ENSDART00000159114
|
znf1129
|
zinc finger protein 1129 |
| chr19_-_5669122 | 0.06 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr4_+_306036 | 0.06 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr20_+_29209926 | 0.06 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr19_+_5480327 | 0.06 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr5_-_12219572 | 0.06 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr1_+_44127292 | 0.05 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr7_+_34453185 | 0.05 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr15_-_22074315 | 0.05 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr8_-_12264486 | 0.05 |
ENSDART00000091612
ENSDART00000135812 |
dab2ipa
|
DAB2 interacting protein a |
| chr1_-_17715493 | 0.05 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr8_-_24332284 | 0.05 |
ENSDART00000163241
|
pimr142
|
Pim proto-oncogene, serine/threonine kinase, related 142 |
| chr7_+_26173751 | 0.05 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr9_+_1313418 | 0.05 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr16_-_32837806 | 0.05 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr8_-_24366726 | 0.05 |
ENSDART00000182351
ENSDART00000187099 ENSDART00000181534 |
CR450805.1
|
|
| chr6_+_7421898 | 0.05 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr2_+_53204750 | 0.04 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr15_+_5360407 | 0.04 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr22_-_20924564 | 0.04 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr11_-_21528056 | 0.04 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_+_16025554 | 0.04 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_-_33824329 | 0.04 |
ENSDART00000189271
ENSDART00000180458 |
vimr2
|
vimentin-related 2 |
| chr11_+_7264457 | 0.04 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr2_+_50608099 | 0.04 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr16_+_54263921 | 0.04 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr8_+_25034544 | 0.04 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_41746427 | 0.04 |
ENSDART00000174232
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr18_+_28106139 | 0.03 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr8_-_16697615 | 0.03 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr23_+_16638639 | 0.03 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr4_+_11723852 | 0.03 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr21_-_33995710 | 0.03 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr15_-_18200358 | 0.03 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr4_-_56954002 | 0.03 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr8_-_39822917 | 0.03 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr13_-_37229245 | 0.03 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr20_+_28803977 | 0.02 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr21_-_45188233 | 0.02 |
ENSDART00000130880
|
si:ch73-269m14.2
|
si:ch73-269m14.2 |
| chr20_+_29209615 | 0.02 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_+_3379673 | 0.02 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr8_-_25034411 | 0.02 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr15_-_33925851 | 0.02 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr20_+_29209767 | 0.01 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr11_+_16152316 | 0.01 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr6_-_40922971 | 0.01 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr18_+_21408794 | 0.01 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr24_-_36680261 | 0.01 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr10_+_35358675 | 0.01 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr25_+_30131055 | 0.01 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr7_+_24023653 | 0.00 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr24_-_18809433 | 0.00 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr24_+_19415124 | 0.00 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx1+rx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |