Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
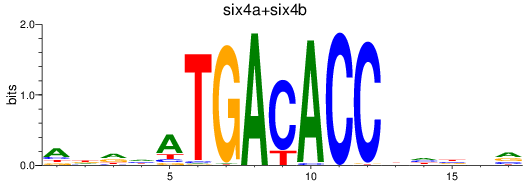
Results for six4a+six4b
Z-value: 0.63
Transcription factors associated with six4a+six4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six4a
|
ENSDARG00000004695 | SIX homeobox 4a |
|
six4b
|
ENSDARG00000031983 | SIX homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six4b | dr11_v1_chr20_+_20484827_20484827 | -0.56 | 3.2e-01 | Click! |
| six4a | dr11_v1_chr13_-_31647323_31647323 | -0.06 | 9.2e-01 | Click! |
Activity profile of six4a+six4b motif
Sorted Z-values of six4a+six4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37837245 | 0.58 |
ENSDART00000171617
|
epd
|
ependymin |
| chr2_+_10134345 | 0.49 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr22_+_25242322 | 0.44 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr22_+_25236888 | 0.40 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr22_+_25236657 | 0.40 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr21_-_26114886 | 0.34 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr22_+_21317597 | 0.34 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr1_-_58562129 | 0.27 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr4_-_22519516 | 0.27 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr9_+_8396755 | 0.27 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr7_-_41964877 | 0.26 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr2_+_50626476 | 0.24 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr4_-_77506362 | 0.24 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr13_-_39399967 | 0.24 |
ENSDART00000190791
ENSDART00000136267 |
slc35f3b
|
solute carrier family 35, member F3b |
| chr1_-_45889820 | 0.23 |
ENSDART00000144735
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr2_+_29491314 | 0.23 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr4_-_14315855 | 0.23 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr12_+_31616412 | 0.22 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr8_+_49149907 | 0.22 |
ENSDART00000138746
|
snpha
|
syntaphilin a |
| chr2_-_30770736 | 0.21 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr11_+_40649412 | 0.21 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr23_-_7125494 | 0.19 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr18_+_7264961 | 0.19 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr7_+_58699900 | 0.19 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr1_-_58561963 | 0.19 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr17_-_13072334 | 0.18 |
ENSDART00000159598
|
CU469462.1
|
|
| chr3_+_12718100 | 0.18 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr21_+_34167178 | 0.17 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr14_-_2209742 | 0.17 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr1_+_37752171 | 0.17 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr18_+_27511976 | 0.17 |
ENSDART00000132017
ENSDART00000140781 |
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr2_+_38804223 | 0.17 |
ENSDART00000147939
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr7_-_54217547 | 0.16 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr3_-_56896702 | 0.16 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr13_-_14269626 | 0.15 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr23_-_33944597 | 0.15 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr16_+_50755133 | 0.14 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr18_-_39188664 | 0.14 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr5_+_13472234 | 0.14 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr21_+_39365920 | 0.14 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr14_-_2318590 | 0.13 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr2_-_14793343 | 0.13 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr2_-_4797512 | 0.13 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr20_+_34717403 | 0.13 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr13_+_3954715 | 0.13 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr7_+_58699718 | 0.13 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr20_+_33534038 | 0.13 |
ENSDART00000029206
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr7_+_13988075 | 0.12 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr17_-_42213285 | 0.12 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr2_-_32558795 | 0.12 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr3_-_40955780 | 0.12 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr17_-_1407593 | 0.12 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr16_-_13818061 | 0.12 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr5_-_43071058 | 0.11 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr10_-_36682509 | 0.11 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr6_+_40591149 | 0.11 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr17_+_7595356 | 0.10 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr17_+_5768608 | 0.10 |
ENSDART00000157039
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr1_-_7894255 | 0.10 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr23_+_27779452 | 0.10 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_48297842 | 0.10 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr9_-_34882516 | 0.10 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr24_-_23839647 | 0.10 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr11_+_41560792 | 0.10 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr2_-_11662851 | 0.08 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr7_-_32599669 | 0.08 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr6_-_35310224 | 0.08 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr23_+_16469530 | 0.08 |
ENSDART00000132898
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr20_+_51813432 | 0.08 |
ENSDART00000127444
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr2_+_33335911 | 0.07 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr23_+_45785563 | 0.07 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr14_+_24042760 | 0.07 |
ENSDART00000106096
|
drd1a
|
dopamine receptor D1a |
| chr17_+_15882533 | 0.06 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr18_-_12416019 | 0.06 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr7_+_6480212 | 0.06 |
ENSDART00000173320
|
si:cabz01036022.1
|
si:cabz01036022.1 |
| chr3_+_311833 | 0.06 |
ENSDART00000187375
|
FP326649.2
|
|
| chr24_-_25166416 | 0.06 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_-_40135942 | 0.05 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr22_+_19262161 | 0.05 |
ENSDART00000163654
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr25_-_1235457 | 0.05 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr1_+_1941031 | 0.05 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr19_+_37135700 | 0.05 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr25_-_17528994 | 0.05 |
ENSDART00000061712
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr6_-_24301324 | 0.05 |
ENSDART00000171401
|
BX927081.1
|
|
| chr19_-_6083761 | 0.05 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr7_+_53498152 | 0.05 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr25_+_19695652 | 0.05 |
ENSDART00000104340
|
mxg
|
myxovirus (influenza virus) resistance G |
| chr15_-_35126332 | 0.04 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr11_+_583725 | 0.04 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr2_+_56213694 | 0.04 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr12_+_1592146 | 0.04 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr6_-_21678263 | 0.04 |
ENSDART00000038777
|
si:dkey-43k4.5
|
si:dkey-43k4.5 |
| chr11_+_24851671 | 0.04 |
ENSDART00000167659
|
ipo9
|
importin 9 |
| chr12_-_18872927 | 0.04 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr2_-_30784198 | 0.04 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr12_+_31422557 | 0.04 |
ENSDART00000153179
|
zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr8_-_7474997 | 0.03 |
ENSDART00000146555
|
gata1b
|
GATA binding protein 1b |
| chr4_-_72468168 | 0.03 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr22_+_15310103 | 0.03 |
ENSDART00000145849
|
CU104797.1
|
|
| chr10_-_32610776 | 0.03 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr20_-_5052786 | 0.03 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr20_-_6184167 | 0.03 |
ENSDART00000147451
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr1_-_46632948 | 0.03 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr17_-_42213822 | 0.02 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr22_-_209741 | 0.02 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr8_-_3346692 | 0.02 |
ENSDART00000057874
|
FUT9 (1 of many)
|
zgc:103510 |
| chr2_-_20666920 | 0.02 |
ENSDART00000143437
ENSDART00000114546 ENSDART00000136113 ENSDART00000179247 |
dusp12
|
dual specificity phosphatase 12 |
| chr5_-_5831037 | 0.02 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr25_-_15214161 | 0.02 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr5_+_66044128 | 0.02 |
ENSDART00000165573
|
jak2b
|
Janus kinase 2b |
| chr9_+_11034314 | 0.02 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr8_+_14058646 | 0.02 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr16_+_11834516 | 0.02 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr14_-_4043818 | 0.01 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr2_+_30465102 | 0.01 |
ENSDART00000188404
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr12_-_3962372 | 0.01 |
ENSDART00000016791
|
eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr7_+_39360797 | 0.01 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr12_-_4475890 | 0.01 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr22_+_2751887 | 0.01 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr3_-_34072446 | 0.01 |
ENSDART00000151528
|
ighv8-2
|
immunoglobulin heavy variable 8-2 |
| chr19_+_16064439 | 0.00 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr21_+_39432248 | 0.00 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr22_+_26853254 | 0.00 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr19_+_770300 | 0.00 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr12_-_31484677 | 0.00 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr15_-_37834433 | 0.00 |
ENSDART00000189748
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr3_-_54607166 | 0.00 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six4a+six4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |