Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
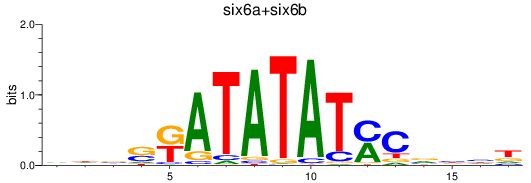
Results for six6a+six6b
Z-value: 0.60
Transcription factors associated with six6a+six6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six6a
|
ENSDARG00000025187 | SIX homeobox 6a |
|
six6b
|
ENSDARG00000031316 | SIX homeobox 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six6a | dr11_v1_chr13_+_31583034_31583034 | 0.60 | 2.8e-01 | Click! |
| six6b | dr11_v1_chr20_-_20533865_20533865 | 0.50 | 4.0e-01 | Click! |
Activity profile of six6a+six6b motif
Sorted Z-values of six6a+six6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_35745572 | 1.13 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr14_+_51098036 | 0.72 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr2_-_44946094 | 0.60 |
ENSDART00000036997
|
camk2n1a
|
calcium/calmodulin-dependent protein kinase II inhibitor 1a |
| chr14_+_50770537 | 0.48 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr9_+_38883388 | 0.41 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr15_-_20939579 | 0.40 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr15_-_25527580 | 0.38 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr2_+_47581997 | 0.36 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_+_1881334 | 0.32 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr13_+_28819768 | 0.31 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr19_+_206835 | 0.31 |
ENSDART00000161137
|
scn1bb
|
sodium channel, voltage-gated, type I, beta b |
| chr19_-_32710922 | 0.31 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr20_-_49729446 | 0.30 |
ENSDART00000111089
|
filip1b
|
filamin A interacting protein 1b |
| chr21_-_26250744 | 0.30 |
ENSDART00000044735
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr21_-_42007213 | 0.28 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr10_+_39199547 | 0.27 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr21_-_26250443 | 0.26 |
ENSDART00000123713
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr7_+_20535869 | 0.25 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr5_-_39736383 | 0.23 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr3_-_39305291 | 0.22 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr5_+_64368770 | 0.22 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr6_+_11026866 | 0.21 |
ENSDART00000189619
|
abcc6a
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6a |
| chr21_-_42055872 | 0.21 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_-_27775236 | 0.20 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr8_-_33114202 | 0.19 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr3_-_18711288 | 0.19 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr3_-_22214029 | 0.19 |
ENSDART00000154818
|
maptb
|
microtubule-associated protein tau b |
| chr24_-_19719240 | 0.19 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr20_+_1960092 | 0.18 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr10_-_38243851 | 0.18 |
ENSDART00000166108
|
usp25
|
ubiquitin specific peptidase 25 |
| chr22_-_12862415 | 0.18 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr12_+_47448558 | 0.17 |
ENSDART00000185689
ENSDART00000105328 |
fmn2b
|
formin 2b |
| chr6_-_49159207 | 0.16 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr16_+_1802307 | 0.16 |
ENSDART00000180026
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr2_+_53357953 | 0.16 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr20_+_12702923 | 0.15 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr2_-_30249746 | 0.15 |
ENSDART00000124153
ENSDART00000142575 |
elocb
|
elongin C paralog b |
| chr1_+_54198893 | 0.15 |
ENSDART00000158948
|
tsc2
|
TSC complex subunit 2 |
| chr15_-_2596125 | 0.15 |
ENSDART00000114923
|
dnajc30b
|
DnaJ (Hsp40) homolog, subfamily C, member 30b |
| chr19_-_3906369 | 0.15 |
ENSDART00000160299
ENSDART00000169205 |
si:ch73-281i18.4
|
si:ch73-281i18.4 |
| chr7_-_31932723 | 0.15 |
ENSDART00000014843
|
bdnf
|
brain-derived neurotrophic factor |
| chr3_-_21106093 | 0.15 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr10_-_5821584 | 0.15 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr5_-_57526807 | 0.14 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr7_-_36358735 | 0.14 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr2_-_144393 | 0.14 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr19_-_3925801 | 0.13 |
ENSDART00000129570
ENSDART00000163138 |
si:ch73-281i18.6
|
si:ch73-281i18.6 |
| chr1_+_27713380 | 0.13 |
ENSDART00000169584
|
si:dkey-29d5.1
|
si:dkey-29d5.1 |
| chr25_+_31277415 | 0.13 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_+_14466860 | 0.13 |
ENSDART00000114097
|
stim2a
|
tromal interaction molecule 2a |
| chr25_+_33072676 | 0.13 |
ENSDART00000182885
ENSDART00000181516 |
tln2b
|
talin 2b |
| chr2_-_30250311 | 0.12 |
ENSDART00000184144
|
elocb
|
elongin C paralog b |
| chr23_-_36934944 | 0.12 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr18_-_14677936 | 0.12 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr6_-_18199062 | 0.12 |
ENSDART00000167513
|
ppp1r27b
|
protein phosphatase 1, regulatory subunit 27b |
| chr18_-_19350792 | 0.12 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr2_-_9607879 | 0.12 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr6_+_28124393 | 0.11 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr18_+_12058403 | 0.11 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr17_+_28882977 | 0.11 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr10_+_45302425 | 0.11 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr10_-_43540196 | 0.11 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr16_+_30002605 | 0.11 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr10_+_7563755 | 0.11 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr16_-_23471387 | 0.11 |
ENSDART00000180899
|
CR533578.1
|
|
| chr10_+_44903676 | 0.10 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr24_+_13017586 | 0.10 |
ENSDART00000142457
|
stau2
|
staufen double-stranded RNA binding protein 2 |
| chr7_+_71955486 | 0.10 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr10_+_23060391 | 0.10 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr12_+_17439580 | 0.10 |
ENSDART00000189257
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr15_-_42736433 | 0.10 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr12_+_2446837 | 0.10 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr20_+_37383034 | 0.09 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr10_+_7564106 | 0.09 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr22_-_10459880 | 0.09 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr25_+_3306620 | 0.09 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr20_+_34596205 | 0.09 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr11_-_17755444 | 0.09 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr18_+_16133595 | 0.09 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr20_-_16849306 | 0.09 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr17_+_24722646 | 0.08 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr23_-_44226556 | 0.08 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr8_+_17143501 | 0.08 |
ENSDART00000061758
|
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr10_-_39198942 | 0.08 |
ENSDART00000077619
|
b3gat1b
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) b |
| chr25_-_24248000 | 0.08 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr18_+_19842569 | 0.07 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr16_+_6632969 | 0.07 |
ENSDART00000109276
|
sall3a
|
spalt-like transcription factor 3a |
| chr13_+_49175624 | 0.07 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr8_-_28357177 | 0.07 |
ENSDART00000182319
|
KLHL12 (1 of many)
|
kelch like family member 12 |
| chr8_-_21142550 | 0.07 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr20_-_40487208 | 0.07 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr7_+_69428831 | 0.07 |
ENSDART00000186975
|
elp5
|
elongator acetyltransferase complex subunit 5 |
| chr20_+_42918755 | 0.07 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr20_-_37287107 | 0.07 |
ENSDART00000076309
|
nmbr
|
neuromedin B receptor |
| chr19_+_27342479 | 0.06 |
ENSDART00000184687
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr24_-_20016817 | 0.06 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr21_+_11105007 | 0.06 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr21_-_35428039 | 0.06 |
ENSDART00000193298
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr16_+_44906324 | 0.06 |
ENSDART00000074960
|
cd22
|
cd22 molecule |
| chr8_-_13184989 | 0.06 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr15_-_31514818 | 0.06 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr2_-_3611960 | 0.06 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr12_-_16380961 | 0.06 |
ENSDART00000086984
|
hectd2
|
HECT domain containing 2 |
| chr17_+_23255365 | 0.06 |
ENSDART00000180277
|
AL935174.5
|
|
| chr18_+_36786842 | 0.06 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr4_+_76426308 | 0.06 |
ENSDART00000180424
ENSDART00000159789 |
FP074874.1
zgc:172128
|
zgc:172128 |
| chr11_-_27702778 | 0.06 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr14_+_45883687 | 0.06 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr2_+_30249977 | 0.06 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr23_+_1702624 | 0.06 |
ENSDART00000149357
|
rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr8_-_37461835 | 0.06 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr23_+_7710447 | 0.05 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr5_-_30079434 | 0.05 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr14_-_2318590 | 0.05 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr14_-_17306261 | 0.05 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr24_-_39772045 | 0.05 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr23_-_38497705 | 0.05 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr4_-_13902188 | 0.05 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr24_-_7587401 | 0.04 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr13_+_46944607 | 0.04 |
ENSDART00000187352
|
fbxo5
|
F-box protein 5 |
| chr18_+_43183749 | 0.04 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr1_-_19233890 | 0.04 |
ENSDART00000127145
|
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr8_-_12847483 | 0.04 |
ENSDART00000146186
|
si:dkey-104n9.1
|
si:dkey-104n9.1 |
| chr13_-_18195942 | 0.04 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr18_+_31410652 | 0.04 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr23_-_24047054 | 0.04 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr11_+_44804685 | 0.04 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr5_+_68267908 | 0.04 |
ENSDART00000130835
|
tnfsf13
|
TNF superfamily member 13 |
| chr3_+_18840810 | 0.04 |
ENSDART00000181137
ENSDART00000128626 ENSDART00000133332 |
tmem104
|
transmembrane protein 104 |
| chr8_+_2575993 | 0.04 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr22_+_19640309 | 0.04 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr10_-_25816558 | 0.03 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr4_+_11458078 | 0.03 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr19_-_17774875 | 0.03 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr4_-_73266929 | 0.03 |
ENSDART00000158194
|
wu:fc30c06
|
wu:fc30c06 |
| chr7_-_7766920 | 0.03 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr12_-_21834611 | 0.03 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr15_-_762319 | 0.03 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr21_-_25756119 | 0.03 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr25_+_1335530 | 0.03 |
ENSDART00000090803
|
fem1b
|
fem-1 homolog b (C. elegans) |
| chr20_-_51946052 | 0.02 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr14_+_26759332 | 0.02 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr3_-_61387273 | 0.02 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr10_+_41549819 | 0.02 |
ENSDART00000114210
|
CABZ01049847.1
|
|
| chr5_+_25304499 | 0.02 |
ENSDART00000163425
|
carnmt1
|
carnosine N-methyltransferase 1 |
| chr14_-_17576391 | 0.02 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr22_-_21845685 | 0.02 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr16_-_36979592 | 0.02 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr5_+_31995683 | 0.02 |
ENSDART00000139744
|
scai
|
suppressor of cancer cell invasion |
| chr20_+_26987416 | 0.02 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr7_+_22313533 | 0.02 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr25_+_15287036 | 0.02 |
ENSDART00000147572
|
hipk3a
|
homeodomain interacting protein kinase 3a |
| chr20_-_29471577 | 0.02 |
ENSDART00000043382
|
grem1a
|
gremlin 1a, DAN family BMP antagonist |
| chr19_+_35002479 | 0.02 |
ENSDART00000103266
|
wdyhv1
|
WDYHV motif containing 1 |
| chr19_+_390298 | 0.01 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr23_+_32335871 | 0.01 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr1_+_31657842 | 0.01 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr7_-_38807819 | 0.01 |
ENSDART00000132772
ENSDART00000052324 |
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr6_-_32315494 | 0.01 |
ENSDART00000127879
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr16_+_3185541 | 0.01 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr7_+_38808027 | 0.01 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr22_+_1431743 | 0.01 |
ENSDART00000182871
|
BX322657.2
|
Danio rerio si:dkeyp-53d3.6 (si:dkeyp-53d3.6), mRNA. |
| chr1_+_34701144 | 0.00 |
ENSDART00000150038
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr15_-_30450898 | 0.00 |
ENSDART00000156584
|
msi2b
|
musashi RNA-binding protein 2b |
| chr13_+_21826096 | 0.00 |
ENSDART00000056432
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr20_-_9580446 | 0.00 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr2_-_23391266 | 0.00 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr20_-_36393555 | 0.00 |
ENSDART00000153421
|
si:dkey-1j5.4
|
si:dkey-1j5.4 |
| chr21_-_2707768 | 0.00 |
ENSDART00000165384
|
CABZ01101739.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of six6a+six6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.3 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.1 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.1 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |