Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
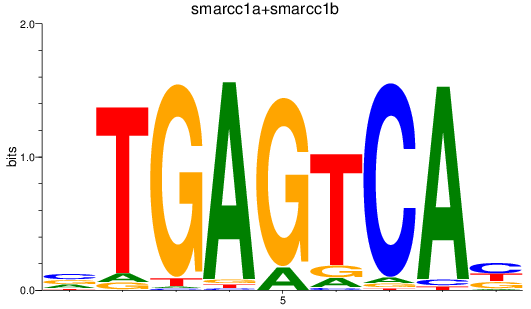
Results for smarcc1a+smarcc1b
Z-value: 1.52
Transcription factors associated with smarcc1a+smarcc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc1a
|
ENSDARG00000017397 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
|
smarcc1b
|
ENSDARG00000098919 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc1b | dr11_v1_chr19_-_19379084_19379084 | -0.53 | 3.6e-01 | Click! |
| smarcc1a | dr11_v1_chr16_-_42461263_42461263 | -0.51 | 3.8e-01 | Click! |
Activity profile of smarcc1a+smarcc1b motif
Sorted Z-values of smarcc1a+smarcc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61185746 | 1.18 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr21_+_25765734 | 0.89 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr21_+_10866421 | 0.84 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr3_-_32818607 | 0.82 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr25_-_22187397 | 0.81 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr17_-_5583345 | 0.79 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr13_-_33134611 | 0.77 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr16_-_22303130 | 0.75 |
ENSDART00000142181
|
si:dkey-92i15.4
|
si:dkey-92i15.4 |
| chr1_-_59252973 | 0.73 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr7_+_7048245 | 0.70 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr3_+_4113551 | 0.70 |
ENSDART00000192309
|
LO018551.1
|
|
| chr22_+_11756040 | 0.70 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr10_+_38593645 | 0.70 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr15_+_46329149 | 0.67 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr16_+_23403602 | 0.66 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr19_-_5332784 | 0.65 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_+_38610741 | 0.65 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr23_+_20408227 | 0.64 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr4_+_76671012 | 0.63 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr19_+_48359259 | 0.61 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr18_+_47313715 | 0.60 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr7_-_20241346 | 0.60 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr13_+_24842857 | 0.58 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr14_+_30279391 | 0.58 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr3_-_4303262 | 0.57 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr13_+_22479988 | 0.57 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr22_-_24858042 | 0.57 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr25_-_13408760 | 0.56 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr25_-_22191733 | 0.54 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr20_+_28364742 | 0.53 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr22_-_24297510 | 0.53 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr18_+_47313899 | 0.52 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr22_+_37888249 | 0.50 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr7_+_17992455 | 0.50 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr2_+_38731696 | 0.49 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr15_-_2657508 | 0.48 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr4_+_76752707 | 0.48 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr14_+_22022441 | 0.48 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr20_+_7084154 | 0.47 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr13_+_22480496 | 0.47 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr8_-_2616326 | 0.47 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr19_-_7420867 | 0.47 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr22_+_11775269 | 0.47 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr2_+_36015049 | 0.47 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr4_+_77948970 | 0.47 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr13_-_7031033 | 0.46 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr15_-_17074393 | 0.44 |
ENSDART00000155526
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr7_+_25033924 | 0.44 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr7_+_34492744 | 0.43 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr10_+_28428222 | 0.42 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr14_-_11456724 | 0.42 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr25_+_10410620 | 0.42 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr19_+_15441022 | 0.41 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_-_1538600 | 0.41 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr16_-_30885838 | 0.40 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr2_-_985417 | 0.40 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr7_-_49892991 | 0.40 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr21_+_45316330 | 0.40 |
ENSDART00000056474
ENSDART00000149314 ENSDART00000149272 ENSDART00000149156 ENSDART00000099497 |
tcf7
|
transcription factor 7 |
| chr1_-_52498146 | 0.40 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr19_+_2670130 | 0.40 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr16_+_18974064 | 0.39 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr9_+_426392 | 0.39 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr4_-_13518381 | 0.39 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr8_+_39663612 | 0.39 |
ENSDART00000188074
|
BX005328.1
|
|
| chr1_-_58913813 | 0.39 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr2_+_3468069 | 0.39 |
ENSDART00000012143
ENSDART00000157116 |
cyp8b3
|
cytochrome P450, family 8, subfamily B, polypeptide 3 |
| chr2_+_3452590 | 0.39 |
ENSDART00000153935
|
cyp8b2
|
cytochrome P450, family 8, subfamily B, polypeptide 2 |
| chr4_+_76787274 | 0.39 |
ENSDART00000156344
|
si:ch73-56d11.3
|
si:ch73-56d11.3 |
| chr8_-_38201415 | 0.39 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr4_+_76659013 | 0.38 |
ENSDART00000147908
ENSDART00000134229 |
ms4a17a.5
|
membrane-spanning 4-domains, subfamily A, member 17A.5 |
| chr3_+_4266289 | 0.37 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr25_-_35524166 | 0.37 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_-_25679339 | 0.37 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr23_-_19500559 | 0.37 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr8_-_23573084 | 0.37 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr1_+_54655160 | 0.37 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr6_+_56141852 | 0.36 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr20_-_26531850 | 0.36 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr22_+_19247255 | 0.36 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr16_-_51288178 | 0.36 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr7_+_35068036 | 0.36 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr17_+_51744450 | 0.36 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr10_-_22095505 | 0.35 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr20_+_2039518 | 0.35 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr5_+_42064144 | 0.35 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr16_+_49005321 | 0.35 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr21_-_7035599 | 0.35 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr20_-_26532167 | 0.35 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr19_-_27570333 | 0.35 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr25_+_29474583 | 0.34 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr8_-_20243389 | 0.34 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr4_+_9508505 | 0.33 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr3_-_27646070 | 0.33 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr8_+_50534948 | 0.33 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr3_-_3939785 | 0.33 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr20_-_35578435 | 0.33 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr13_+_23988442 | 0.33 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_+_4346854 | 0.32 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr23_-_39636195 | 0.32 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr1_+_6640437 | 0.32 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr16_+_26547152 | 0.32 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr10_-_44560165 | 0.32 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr7_+_19482084 | 0.32 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr1_-_10071422 | 0.32 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr16_-_14397003 | 0.31 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr1_-_35694978 | 0.31 |
ENSDART00000136157
|
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr8_-_18667693 | 0.31 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr16_-_12487657 | 0.31 |
ENSDART00000189863
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr2_-_3437862 | 0.31 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr25_+_13205878 | 0.31 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr7_-_71389375 | 0.31 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr17_-_30635298 | 0.31 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr3_-_4760384 | 0.30 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr14_-_40821411 | 0.30 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr8_+_6576940 | 0.30 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr23_+_24272421 | 0.30 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr10_-_44482911 | 0.30 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr3_-_48259289 | 0.30 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr19_+_7895086 | 0.30 |
ENSDART00000132180
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr13_-_16222388 | 0.30 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr3_-_58831683 | 0.29 |
ENSDART00000110292
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr16_-_12491901 | 0.29 |
ENSDART00000174467
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr10_-_8079737 | 0.29 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr12_+_22576404 | 0.29 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr16_-_17200120 | 0.29 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr16_+_20915319 | 0.29 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr16_-_16522013 | 0.29 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr4_+_76775837 | 0.29 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr17_-_53359028 | 0.28 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr6_-_8244474 | 0.28 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr24_+_13869092 | 0.28 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_-_13818061 | 0.28 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr9_+_56422311 | 0.28 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr16_+_21790870 | 0.28 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr24_+_38201089 | 0.28 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr20_+_34770197 | 0.28 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr16_-_36834505 | 0.28 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr6_+_51713076 | 0.28 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr8_-_11170114 | 0.27 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr2_+_25278107 | 0.27 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr14_+_30774515 | 0.27 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr13_+_4671698 | 0.27 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr8_+_13389115 | 0.27 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr23_+_25856541 | 0.27 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr15_-_38129845 | 0.27 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr16_-_12496632 | 0.27 |
ENSDART00000019941
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr8_+_8671229 | 0.27 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr22_+_19528851 | 0.27 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr8_-_40075983 | 0.26 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr12_-_28363111 | 0.26 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr4_+_77933084 | 0.26 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_-_12198765 | 0.26 |
ENSDART00000104203
ENSDART00000128364 ENSDART00000166887 ENSDART00000041533 |
krt95
|
kertain 95 |
| chr7_+_34549377 | 0.26 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr19_-_703898 | 0.26 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr22_-_13350240 | 0.26 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr23_+_25232711 | 0.26 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr22_-_26595027 | 0.26 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr6_+_40723554 | 0.26 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr1_+_59088205 | 0.26 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr1_-_52497834 | 0.26 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr23_-_36316352 | 0.26 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr8_-_20291922 | 0.26 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr4_-_77216726 | 0.25 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr11_-_21014572 | 0.25 |
ENSDART00000042220
|
slc6a14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr5_-_25582721 | 0.25 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr1_+_57235896 | 0.25 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr19_-_27564458 | 0.25 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr5_+_6670945 | 0.25 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr12_-_6159545 | 0.25 |
ENSDART00000152487
|
rltgr
|
RAMP-like triterpene glycoside receptor |
| chr7_+_15736230 | 0.25 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr21_-_11657043 | 0.24 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr14_+_30774894 | 0.24 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr9_-_9282519 | 0.24 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr20_-_13660600 | 0.24 |
ENSDART00000063826
|
TAGAP
|
si:ch211-122h15.4 |
| chr22_+_19188809 | 0.24 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr7_+_20512419 | 0.24 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr6_+_52947186 | 0.24 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr12_-_37299646 | 0.24 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr4_+_5317483 | 0.23 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr3_-_32859335 | 0.23 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr1_+_14253118 | 0.23 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr14_+_30774032 | 0.23 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr20_+_35445462 | 0.23 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr25_+_29474982 | 0.23 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr11_+_1551603 | 0.23 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr24_+_38208823 | 0.23 |
ENSDART00000187538
ENSDART00000190189 |
CU896602.3
|
|
| chr23_-_4975452 | 0.23 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr20_-_13660767 | 0.23 |
ENSDART00000127654
|
TAGAP
|
si:ch211-122h15.4 |
| chr3_+_31680592 | 0.23 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr5_-_1047504 | 0.23 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr5_-_30615901 | 0.23 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr23_+_43718115 | 0.23 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr13_-_15982707 | 0.23 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr21_-_42876565 | 0.23 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr25_+_23336310 | 0.22 |
ENSDART00000156457
|
ptprjb.2
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 2 |
| chr5_+_19337108 | 0.22 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr8_+_28695914 | 0.22 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr9_-_11551608 | 0.22 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr25_-_29072162 | 0.22 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr6_+_56147812 | 0.22 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of smarcc1a+smarcc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.8 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.4 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.2 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.7 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.2 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.2 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.8 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 0.2 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0048914 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.3 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.3 | GO:0006925 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) myoblast migration involved in skeletal muscle regeneration(GO:0014839) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) myoblast migration(GO:0051451) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.2 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 1.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0046436 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.3 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.9 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.0 | GO:0051230 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.0 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.1 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 2.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.2 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 1.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.3 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 1.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |