Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
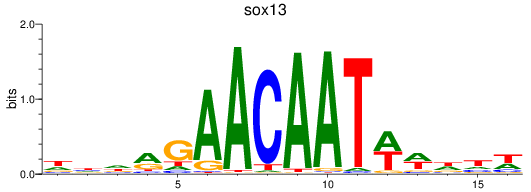
Results for sox13
Z-value: 0.55
Transcription factors associated with sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox13
|
ENSDARG00000030297 | SRY-box transcription factor 13 |
|
sox13
|
ENSDARG00000112638 | SRY-box transcription factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox13 | dr11_v1_chr11_+_37768298_37768298 | -0.93 | 2.0e-02 | Click! |
Activity profile of sox13 motif
Sorted Z-values of sox13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_15602760 | 0.43 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr16_-_17197546 | 0.28 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr21_+_5589923 | 0.27 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr21_+_40092301 | 0.26 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr12_+_47663419 | 0.26 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr21_+_38817785 | 0.25 |
ENSDART00000177616
ENSDART00000149085 |
hnf1bb
|
HNF1 homeobox Bb |
| chr7_+_25033924 | 0.23 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr11_-_29657947 | 0.23 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr22_-_15602593 | 0.22 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr20_+_34029820 | 0.22 |
ENSDART00000143901
ENSDART00000134305 |
prg4b
|
proteoglycan 4b |
| chr1_-_19079957 | 0.19 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr15_-_21155641 | 0.19 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr11_+_29671661 | 0.19 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr6_-_42003780 | 0.19 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr9_-_16109001 | 0.18 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr6_-_39765546 | 0.18 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr1_+_12231478 | 0.17 |
ENSDART00000111485
|
tmod1
|
tropomodulin 1 |
| chr17_-_6382392 | 0.17 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr24_-_2381143 | 0.17 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr13_+_50375800 | 0.17 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr20_+_10538025 | 0.17 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr14_-_36409960 | 0.17 |
ENSDART00000173140
|
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr21_-_32119965 | 0.17 |
ENSDART00000185015
ENSDART00000123419 ENSDART00000193026 |
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr9_+_50316921 | 0.16 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr9_-_12443726 | 0.16 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr24_+_39105051 | 0.16 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr7_-_55648336 | 0.16 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr23_+_31815423 | 0.16 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr18_-_49286381 | 0.16 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr3_-_44059902 | 0.16 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr8_+_17069577 | 0.16 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr7_-_29292206 | 0.15 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr2_+_52232630 | 0.15 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr12_+_19191787 | 0.15 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr2_+_16780643 | 0.15 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr23_-_42232124 | 0.15 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr14_+_5383060 | 0.14 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr7_+_65673885 | 0.14 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr8_+_30664077 | 0.14 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr15_-_33965440 | 0.14 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr20_-_33583779 | 0.14 |
ENSDART00000097823
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr11_+_16216909 | 0.14 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr12_+_30586599 | 0.14 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr7_-_51757085 | 0.14 |
ENSDART00000167747
|
leap2
|
liver-expressed antimicrobial peptide 2 |
| chr22_+_1887750 | 0.14 |
ENSDART00000180917
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr24_+_22485710 | 0.14 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr18_-_38270077 | 0.13 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr12_-_6177894 | 0.13 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr7_-_51953807 | 0.13 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr11_-_23458792 | 0.13 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr20_+_34845997 | 0.13 |
ENSDART00000035612
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr2_+_20406399 | 0.13 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr14_+_16151636 | 0.13 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr16_-_17072440 | 0.13 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr20_-_35076387 | 0.13 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr10_-_1276046 | 0.13 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr2_+_3502430 | 0.13 |
ENSDART00000113395
|
SERP1
|
zgc:92744 |
| chr4_-_10599062 | 0.13 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr7_-_32980017 | 0.13 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr15_-_33964897 | 0.13 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr25_+_22320738 | 0.13 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr23_+_42810055 | 0.12 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr11_-_10643091 | 0.12 |
ENSDART00000192314
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr6_-_58764672 | 0.12 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr14_+_16151368 | 0.12 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr7_+_20467549 | 0.12 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr14_+_30285613 | 0.12 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr21_-_2261720 | 0.12 |
ENSDART00000170161
|
si:ch73-299h12.2
|
si:ch73-299h12.2 |
| chr19_+_24575077 | 0.12 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr22_-_25612680 | 0.12 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr14_+_26247319 | 0.12 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr13_-_17723417 | 0.12 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr23_+_31107685 | 0.12 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr3_-_42967698 | 0.12 |
ENSDART00000187938
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr7_+_60079302 | 0.12 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr23_-_35195908 | 0.12 |
ENSDART00000122429
|
klf15
|
Kruppel-like factor 15 |
| chr8_-_28304534 | 0.12 |
ENSDART00000078545
|
lmod1a
|
leiomodin 1a (smooth muscle) |
| chr16_+_13993285 | 0.11 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr14_+_15331486 | 0.11 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr10_-_21545091 | 0.11 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr25_-_35124576 | 0.11 |
ENSDART00000128017
|
zgc:171759
|
zgc:171759 |
| chr1_+_36665555 | 0.11 |
ENSDART00000128557
ENSDART00000010632 |
ednraa
|
endothelin receptor type Aa |
| chr25_-_12923482 | 0.11 |
ENSDART00000161754
|
CR450808.1
|
|
| chr24_-_38083378 | 0.11 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr7_-_52096498 | 0.11 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr19_-_1947403 | 0.11 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr25_-_3830272 | 0.11 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr3_-_8246309 | 0.11 |
ENSDART00000134497
ENSDART00000015232 |
cyp2k22
|
cytochrome P450, family 2, subfamily K, polypeptide 22 |
| chr16_+_44298902 | 0.11 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr2_-_5071888 | 0.11 |
ENSDART00000185036
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr6_-_52675630 | 0.11 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr22_-_28624879 | 0.11 |
ENSDART00000188437
|
adgrg7.2
|
adhesion G protein-coupled receptor G7, tandem duplicate 2 |
| chr21_-_21828261 | 0.11 |
ENSDART00000151181
|
defbl2
|
defensin, beta-like 2 |
| chr20_-_40367493 | 0.11 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr21_+_15833222 | 0.11 |
ENSDART00000151161
|
si:dkeyp-87d8.8
|
si:dkeyp-87d8.8 |
| chr2_+_37875789 | 0.10 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr8_-_32497581 | 0.10 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr6_+_8626427 | 0.10 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr22_-_17606575 | 0.10 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr9_+_22359919 | 0.10 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr2_-_27329667 | 0.10 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr14_+_15430991 | 0.10 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr15_+_22394074 | 0.10 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr2_-_51259371 | 0.10 |
ENSDART00000158049
ENSDART00000164916 ENSDART00000157792 |
si:ch211-215e19.6
|
si:ch211-215e19.6 |
| chr3_+_12835370 | 0.10 |
ENSDART00000166089
|
si:ch211-8c17.3
|
si:ch211-8c17.3 |
| chr6_-_7052408 | 0.10 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr5_-_9216758 | 0.10 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr5_+_9417409 | 0.10 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr14_-_46198373 | 0.10 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr6_+_39905021 | 0.10 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr13_-_40398556 | 0.10 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr20_-_14054083 | 0.10 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr23_+_42819221 | 0.10 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr8_-_32497815 | 0.09 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr15_+_45643787 | 0.09 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr22_+_10651726 | 0.09 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr8_+_40476811 | 0.09 |
ENSDART00000129772
|
gck
|
glucokinase (hexokinase 4) |
| chr13_+_35690023 | 0.09 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr7_-_35516251 | 0.09 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr2_+_20605925 | 0.09 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr18_-_10713230 | 0.09 |
ENSDART00000183646
|
rbm28
|
RNA binding motif protein 28 |
| chr4_+_69150355 | 0.09 |
ENSDART00000172364
ENSDART00000184374 |
si:ch211-209j12.2
|
si:ch211-209j12.2 |
| chr11_+_29965822 | 0.09 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr9_-_23944470 | 0.09 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr15_+_31515976 | 0.09 |
ENSDART00000156471
|
tex26
|
testis expressed 26 |
| chr10_+_35439952 | 0.09 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr22_-_38274995 | 0.09 |
ENSDART00000179786
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr16_-_563732 | 0.09 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr18_+_45781516 | 0.09 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr2_+_37207461 | 0.09 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr2_-_23479714 | 0.09 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr17_-_45386546 | 0.09 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr21_+_19062124 | 0.09 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr22_+_3153876 | 0.09 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr7_+_17393794 | 0.09 |
ENSDART00000059673
|
nitr7b
|
novel immune-type receptor 7b |
| chr9_-_21498880 | 0.09 |
ENSDART00000131634
|
aff3
|
AF4/FMR2 family, member 3 |
| chr13_+_38213694 | 0.09 |
ENSDART00000057320
|
zgc:171579
|
zgc:171579 |
| chr7_+_17992640 | 0.09 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr20_-_27225876 | 0.09 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr9_+_32931625 | 0.09 |
ENSDART00000143484
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr14_-_11456724 | 0.09 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr5_+_44944778 | 0.09 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr9_+_1039215 | 0.09 |
ENSDART00000192186
|
DOCK9 (1 of many)
|
dedicator of cytokinesis 9 |
| chr7_-_20464468 | 0.09 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr4_-_55592720 | 0.09 |
ENSDART00000164747
|
znf1029
|
zinc finger protein 1029 |
| chr6_-_7052595 | 0.09 |
ENSDART00000081761
ENSDART00000181351 |
bin1b
|
bridging integrator 1b |
| chr11_-_25539323 | 0.09 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr10_-_21362071 | 0.09 |
ENSDART00000125167
|
avd
|
avidin |
| chr7_-_41512999 | 0.09 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr5_+_46196147 | 0.09 |
ENSDART00000084112
|
f2r
|
coagulation factor II (thrombin) receptor |
| chr14_-_25599002 | 0.09 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr16_-_42014272 | 0.08 |
ENSDART00000180488
|
etv2
|
ets variant 2 |
| chr16_-_21853082 | 0.08 |
ENSDART00000166677
|
si:ch73-34h11.1
|
si:ch73-34h11.1 |
| chr1_-_55262763 | 0.08 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr3_+_32689707 | 0.08 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr4_+_72578548 | 0.08 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr18_-_17399291 | 0.08 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr4_-_16330368 | 0.08 |
ENSDART00000128932
|
epyc
|
epiphycan |
| chr7_+_17992455 | 0.08 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr7_-_35515931 | 0.08 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr2_+_55365727 | 0.08 |
ENSDART00000162943
|
FP245456.1
|
|
| chr19_-_35428815 | 0.08 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr13_+_35689749 | 0.08 |
ENSDART00000158726
|
psme4a
|
proteasome activator subunit 4a |
| chr6_-_39275793 | 0.08 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr2_-_23443189 | 0.08 |
ENSDART00000168894
|
si:ch211-14p21.4
|
si:ch211-14p21.4 |
| chr3_+_26081343 | 0.08 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr2_+_20410652 | 0.08 |
ENSDART00000185940
|
palmda
|
palmdelphin a |
| chr5_+_27434601 | 0.08 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr8_-_41279326 | 0.08 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr22_+_883678 | 0.08 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr5_+_1983495 | 0.08 |
ENSDART00000179860
|
CT573103.2
|
|
| chr22_+_9287929 | 0.08 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr11_-_42752884 | 0.08 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr10_+_39263827 | 0.08 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr25_+_5972690 | 0.08 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr2_+_20605186 | 0.08 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr13_-_31397987 | 0.08 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr13_-_23074238 | 0.08 |
ENSDART00000143210
|
srgn
|
serglycin |
| chr3_-_8272632 | 0.08 |
ENSDART00000143522
|
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr10_-_21362320 | 0.08 |
ENSDART00000189789
|
avd
|
avidin |
| chr22_-_12746539 | 0.08 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr7_+_3229828 | 0.08 |
ENSDART00000135367
|
CR855393.2
|
|
| chr22_+_6198742 | 0.08 |
ENSDART00000132889
|
si:dkey-19a16.11
|
si:dkey-19a16.11 |
| chr7_+_3125746 | 0.08 |
ENSDART00000173182
|
si:ch73-122k23.1
|
si:ch73-122k23.1 |
| chr14_+_30775066 | 0.08 |
ENSDART00000139975
ENSDART00000144921 |
atl3
|
atlastin 3 |
| chr20_+_35438300 | 0.08 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr22_+_38192568 | 0.08 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr24_+_17270129 | 0.08 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr19_-_24555623 | 0.08 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr21_-_22720381 | 0.08 |
ENSDART00000130789
|
c1qc
|
complement component 1, q subcomponent, C chain |
| chr3_-_57744323 | 0.08 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr24_-_11446156 | 0.07 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr8_+_22359881 | 0.07 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr25_+_21046856 | 0.07 |
ENSDART00000181343
|
chrm2b
|
cholinergic receptor, muscarinic 2b |
| chr21_-_21530868 | 0.07 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr9_+_22657221 | 0.07 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr17_+_4207278 | 0.07 |
ENSDART00000193612
|
CU929304.1
|
|
| chr22_-_22337382 | 0.07 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr13_+_28417297 | 0.07 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr6_-_23931442 | 0.07 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.2 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.0 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.3 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.1 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.0 | 0.2 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |