Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
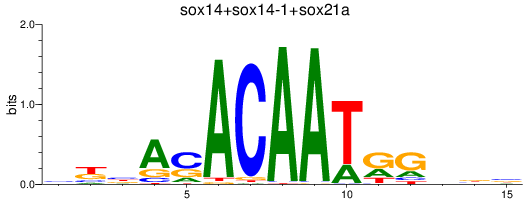
Results for sox14+sox14-1+sox21a
Z-value: 1.26
Transcription factors associated with sox14+sox14-1+sox21a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox21a
|
ENSDARG00000031664 | SRY-box transcription factor 21a |
|
sox14
|
ENSDARG00000070929 | SRY-box transcription factor 14 |
|
sox21a
|
ENSDARG00000111842 | SRY-box transcription factor 21a |
|
sox14-1
|
ENSDARG00000111969 | SRY-box transcription factor 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FQ377628.2 | dr11_v1_chr2_-_22575851_22575851 | -0.80 | 1.1e-01 | Click! |
| sox14 | dr11_v1_chr6_-_26559921_26559921 | -0.70 | 1.9e-01 | Click! |
| sox21a | dr11_v1_chr6_+_7414215_7414215 | 0.25 | 6.8e-01 | Click! |
Activity profile of sox14+sox14-1+sox21a motif
Sorted Z-values of sox14+sox14-1+sox21a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_11457500 | 1.51 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr25_+_25202213 | 1.40 |
ENSDART00000150744
|
ccl39a.10
|
chemokine (C-C motif) ligand 39, duplicate 10 |
| chr17_+_53292215 | 1.19 |
ENSDART00000170686
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr24_-_11076400 | 1.12 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr15_-_21132480 | 1.07 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr12_-_4346085 | 1.04 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr25_-_12982193 | 0.99 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr19_-_7420867 | 0.90 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr23_-_3758637 | 0.89 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr12_-_17655683 | 0.87 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr20_+_54312970 | 0.85 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr2_-_39017838 | 0.81 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr22_-_15693085 | 0.74 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr5_+_43782267 | 0.73 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr3_-_34069637 | 0.73 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr18_+_46151505 | 0.71 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr8_-_32385989 | 0.70 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr5_-_41531629 | 0.69 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr2_-_36933472 | 0.69 |
ENSDART00000170405
|
BX470229.2
|
|
| chr16_-_51888952 | 0.69 |
ENSDART00000186407
ENSDART00000175435 |
rpl28
|
ribosomal protein L28 |
| chr8_-_45760087 | 0.68 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr20_+_54309148 | 0.66 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr23_-_6641223 | 0.66 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr20_+_54295213 | 0.65 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr20_+_54299419 | 0.65 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr22_-_24967348 | 0.64 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr15_+_22390076 | 0.64 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr1_-_49225890 | 0.63 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr14_+_46313135 | 0.63 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_+_37068223 | 0.59 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr2_-_56348727 | 0.59 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr12_-_2800809 | 0.59 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr9_-_24244383 | 0.58 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr9_+_563547 | 0.57 |
ENSDART00000162761
|
CU984600.2
|
|
| chr1_-_18848955 | 0.57 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr14_-_246342 | 0.56 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr3_-_59297532 | 0.55 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr6_+_7466223 | 0.55 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr6_+_584632 | 0.55 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr16_-_17056630 | 0.53 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr12_+_22576404 | 0.53 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_-_50139860 | 0.51 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr23_-_19715799 | 0.51 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr10_-_342564 | 0.50 |
ENSDART00000157633
|
vaspa
|
vasodilator stimulated phosphoprotein a |
| chr8_-_46486009 | 0.49 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr2_+_36004381 | 0.48 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr6_+_30668098 | 0.47 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr3_+_3545825 | 0.47 |
ENSDART00000109060
|
CR589947.1
|
|
| chr8_-_20243389 | 0.46 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr25_-_13023569 | 0.45 |
ENSDART00000167232
|
ccl39.1
|
chemokine (C-C motif) ligand 39, duplicate 1 |
| chr2_-_28420415 | 0.44 |
ENSDART00000183857
|
CABZ01056051.1
|
|
| chr22_+_30631072 | 0.43 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr1_+_59088205 | 0.43 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr11_+_14280598 | 0.42 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr7_-_19915414 | 0.42 |
ENSDART00000144180
|
zgc:110591
|
zgc:110591 |
| chr7_-_51953807 | 0.41 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr3_+_41714966 | 0.40 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr11_-_11791718 | 0.40 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr16_-_40373836 | 0.39 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr5_-_30615901 | 0.39 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr15_+_29025090 | 0.39 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr15_+_37331585 | 0.39 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr7_-_34294613 | 0.39 |
ENSDART00000191632
|
CR388195.1
|
|
| chr11_-_44498975 | 0.38 |
ENSDART00000173066
ENSDART00000189976 |
si:ch1073-365p7.2
|
si:ch1073-365p7.2 |
| chr5_+_38680247 | 0.37 |
ENSDART00000132763
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr5_+_38631687 | 0.37 |
ENSDART00000147280
|
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr15_+_38006366 | 0.37 |
ENSDART00000156162
|
si:dkey-238d18.13
|
si:dkey-238d18.13 |
| chr15_-_36533322 | 0.36 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr7_+_35268880 | 0.36 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr11_+_29965822 | 0.35 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr25_-_12998202 | 0.35 |
ENSDART00000171661
|
ccl39.3
|
chemokine (C-C motif) ligand 39, duplicate 3 |
| chr19_-_32849036 | 0.35 |
ENSDART00000131518
|
si:dkey-284p5.3
|
si:dkey-284p5.3 |
| chr16_+_35535375 | 0.35 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr23_+_10352921 | 0.35 |
ENSDART00000081193
|
KRT18 (1 of many)
|
si:ch211-133j6.3 |
| chr2_+_36608387 | 0.35 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr4_+_69262318 | 0.35 |
ENSDART00000188841
|
si:ch211-209j12.6
|
si:ch211-209j12.6 |
| chr21_-_44081540 | 0.34 |
ENSDART00000130833
|
FO704810.1
|
|
| chr17_+_14711765 | 0.34 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr8_-_46457233 | 0.34 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr1_-_56717813 | 0.34 |
ENSDART00000159196
|
FO681314.1
|
|
| chr17_-_6600899 | 0.33 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr19_-_5125663 | 0.33 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr5_-_26181863 | 0.32 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr14_+_46313396 | 0.31 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_-_34054081 | 0.31 |
ENSDART00000151590
|
ighv1-2
|
immunoglobulin heavy variable 1-2 |
| chr17_-_6599484 | 0.31 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr6_+_17789481 | 0.31 |
ENSDART00000190768
|
CR376861.1
|
|
| chr12_+_19030391 | 0.30 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr24_-_26485098 | 0.30 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr14_+_26439227 | 0.30 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr2_-_45510699 | 0.30 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr16_+_5926520 | 0.29 |
ENSDART00000162229
|
ulk4
|
unc-51 like kinase 4 |
| chr17_-_44402713 | 0.29 |
ENSDART00000190343
|
BX548045.2
|
|
| chr7_+_38380135 | 0.28 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr17_-_20228610 | 0.28 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr8_-_23884301 | 0.28 |
ENSDART00000185509
ENSDART00000147202 |
lhfpl5b
|
LHFPL tetraspan subfamily member 5b |
| chr8_-_30204650 | 0.28 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr1_-_55118745 | 0.28 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr14_-_38872536 | 0.28 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr6_+_12968101 | 0.27 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr11_+_36665359 | 0.27 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr4_-_43070566 | 0.27 |
ENSDART00000183779
|
si:dkey-54j5.2
|
si:dkey-54j5.2 |
| chr24_-_26484298 | 0.27 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr19_+_43716607 | 0.27 |
ENSDART00000133199
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr10_+_32662886 | 0.26 |
ENSDART00000099841
|
rad51c
|
RAD51 paralog C |
| chr24_+_13642126 | 0.26 |
ENSDART00000126769
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr7_+_8324506 | 0.25 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr3_+_53240562 | 0.25 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr3_-_34068044 | 0.25 |
ENSDART00000187926
|
ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr1_+_56755536 | 0.25 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr5_-_57686487 | 0.25 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr16_+_35535171 | 0.24 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr25_+_27405738 | 0.24 |
ENSDART00000183266
ENSDART00000115139 |
pot1
|
protection of telomeres 1 homolog |
| chr24_-_21914276 | 0.24 |
ENSDART00000128687
|
c1qtnf9
|
C1q and TNF related 9 |
| chr12_-_33770299 | 0.24 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr6_+_43426599 | 0.24 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr20_+_33922339 | 0.24 |
ENSDART00000019059
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr13_+_30912938 | 0.23 |
ENSDART00000190003
|
drgx
|
dorsal root ganglia homeobox |
| chr4_+_5392094 | 0.23 |
ENSDART00000135122
|
si:dkey-14d8.20
|
si:dkey-14d8.20 |
| chr15_+_38071874 | 0.23 |
ENSDART00000125770
|
si:ch73-380l3.5
|
si:ch73-380l3.5 |
| chr20_+_35445462 | 0.23 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr16_-_21799407 | 0.23 |
ENSDART00000123717
|
FP015862.4
|
|
| chr22_+_4443689 | 0.23 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr2_-_51500957 | 0.23 |
ENSDART00000172481
|
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr19_-_11949996 | 0.23 |
ENSDART00000163478
ENSDART00000167299 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr22_+_24770744 | 0.23 |
ENSDART00000142882
|
si:rp71-23d18.4
|
si:rp71-23d18.4 |
| chr9_+_21243985 | 0.23 |
ENSDART00000131586
|
si:rp71-68n21.12
|
si:rp71-68n21.12 |
| chr13_-_37636147 | 0.22 |
ENSDART00000144065
|
si:dkey-188i13.8
|
si:dkey-188i13.8 |
| chr4_+_39055027 | 0.22 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr1_+_218524 | 0.22 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr2_-_51512294 | 0.22 |
ENSDART00000185287
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr13_-_36761379 | 0.22 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr10_+_33171501 | 0.22 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr9_-_21838045 | 0.22 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr6_-_25371196 | 0.21 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr7_+_9922607 | 0.21 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr3_-_10751491 | 0.21 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr4_-_22749553 | 0.21 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr16_-_24703603 | 0.20 |
ENSDART00000131167
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr19_+_43715911 | 0.19 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr15_+_31471808 | 0.19 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr2_+_43864575 | 0.19 |
ENSDART00000151951
|
nlrb5
|
NOD-like receptor family B, member 5 |
| chr20_-_7547080 | 0.19 |
ENSDART00000146135
|
usp24
|
ubiquitin specific peptidase 24 |
| chr7_+_67381912 | 0.19 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr9_-_2936017 | 0.19 |
ENSDART00000102823
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr5_+_38743644 | 0.19 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr17_-_32863250 | 0.19 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr16_+_42464613 | 0.19 |
ENSDART00000162454
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr5_-_23715027 | 0.19 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr1_-_35695614 | 0.19 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr12_+_17865374 | 0.18 |
ENSDART00000169019
|
tmem130
|
transmembrane protein 130 |
| chr23_-_35491587 | 0.18 |
ENSDART00000153794
|
fam110c
|
family with sequence similarity 110, member C |
| chr14_+_9462726 | 0.18 |
ENSDART00000063559
|
rell2
|
RELT-like 2 |
| chr20_+_27749133 | 0.18 |
ENSDART00000089013
|
vrtn
|
vertebrae development associated |
| chr21_+_12056934 | 0.18 |
ENSDART00000125380
|
zgc:162344
|
zgc:162344 |
| chr14_+_16036139 | 0.18 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr13_-_38088627 | 0.18 |
ENSDART00000175268
|
CT027676.1
|
|
| chr5_-_26834511 | 0.18 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr5_+_38680061 | 0.18 |
ENSDART00000143259
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr16_-_32204470 | 0.18 |
ENSDART00000143928
|
rwdd1
|
RWD domain containing 1 |
| chr3_-_5644028 | 0.17 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr13_+_30912385 | 0.17 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr14_-_970853 | 0.17 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr19_-_41404870 | 0.17 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr11_+_16138065 | 0.17 |
ENSDART00000188616
|
pimr204
|
Pim proto-oncogene, serine/threonine kinase, related 204 |
| chr6_-_39765546 | 0.17 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr17_+_2727807 | 0.16 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr21_-_16113477 | 0.16 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr16_-_24703855 | 0.16 |
ENSDART00000182422
|
negaly6
|
neuromast-expressed gpi-anchored lymphocyte antigen 6 |
| chr18_+_20034023 | 0.16 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr16_-_41488023 | 0.16 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr16_-_41487589 | 0.16 |
ENSDART00000188115
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr4_-_41209037 | 0.16 |
ENSDART00000151948
|
si:ch211-73m21.1
|
si:ch211-73m21.1 |
| chr9_-_11560110 | 0.16 |
ENSDART00000176941
|
cryba2b
|
crystallin, beta A2b |
| chr21_-_16113799 | 0.16 |
ENSDART00000187848
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr3_+_52078798 | 0.16 |
ENSDART00000156882
|
si:dkey-88e12.3
|
si:dkey-88e12.3 |
| chr21_+_21621042 | 0.16 |
ENSDART00000134907
|
tgfb1b
|
transforming growth factor, beta 1b |
| chr14_+_30910114 | 0.15 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr3_-_18675688 | 0.15 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr25_-_16160494 | 0.15 |
ENSDART00000146880
|
si:dkey-80c24.4
|
si:dkey-80c24.4 |
| chr6_-_1591002 | 0.15 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr4_-_71110826 | 0.15 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr16_+_26846495 | 0.15 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr15_+_11644866 | 0.15 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr20_+_9128256 | 0.14 |
ENSDART00000163883
ENSDART00000183072 ENSDART00000187276 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr25_+_35058088 | 0.14 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr12_-_44010532 | 0.14 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr18_-_33254039 | 0.14 |
ENSDART00000099130
|
si:ch211-229c8.4
|
si:ch211-229c8.4 |
| chr13_+_8677166 | 0.14 |
ENSDART00000181016
ENSDART00000135028 |
prop1
|
PROP paired-like homeobox 1 |
| chr12_-_17479078 | 0.13 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr2_-_6112862 | 0.13 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr15_-_37779978 | 0.13 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr2_+_36939749 | 0.13 |
ENSDART00000005382
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr11_+_583725 | 0.13 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr25_-_15268443 | 0.12 |
ENSDART00000151827
|
ccdc73
|
coiled-coil domain containing 73 |
| chr4_-_36139585 | 0.12 |
ENSDART00000132071
|
znf992
|
zinc finger protein 992 |
| chr9_-_11560427 | 0.12 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr12_-_4916969 | 0.12 |
ENSDART00000161680
|
ifnphi4
|
interferon phi 4 |
| chr12_-_5998898 | 0.12 |
ENSDART00000142659
ENSDART00000004896 |
kat7b
|
K(lysine) acetyltransferase 7b |
| chr4_+_35901451 | 0.12 |
ENSDART00000131142
|
znf1088
|
zinc finger protein 1088 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox14+sox14-1+sox21a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0044878 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.2 | 0.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.7 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.6 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 3.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.3 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.2 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.0 | 0.5 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0030801 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of cyclase activity(GO:0031281) positive regulation of adenylate cyclase activity(GO:0045762) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.0 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 2.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0071459 | mitotic metaphase plate congression(GO:0007080) protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.5 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |