Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
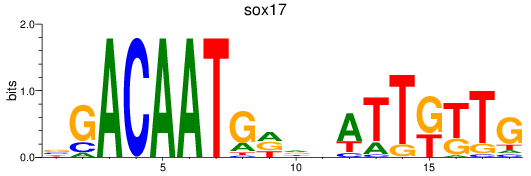
Results for sox17
Z-value: 0.95
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox17 | dr11_v1_chr7_-_58776400_58776400 | 0.24 | 7.0e-01 | Click! |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_11493236 | 0.64 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr4_-_14328997 | 0.48 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr5_-_20678300 | 0.42 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr24_-_38079261 | 0.41 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr1_-_12278056 | 0.39 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr1_-_42779075 | 0.39 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr10_+_4235998 | 0.38 |
ENSDART00000169328
|
plppr1
|
phospholipid phosphatase related 1 |
| chr15_-_33896159 | 0.36 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr2_+_3472832 | 0.33 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr10_-_2522588 | 0.33 |
ENSDART00000081926
|
CU856539.1
|
|
| chr25_-_1720736 | 0.33 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr20_+_40457599 | 0.32 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr2_-_12502495 | 0.31 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr16_-_28658341 | 0.31 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr11_+_35364445 | 0.31 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr1_+_44800298 | 0.31 |
ENSDART00000073712
ENSDART00000187054 |
tmem187
|
transmembrane protein 187 |
| chr8_+_6967108 | 0.31 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr2_+_26240631 | 0.30 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr5_-_26323137 | 0.30 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr21_+_33503835 | 0.30 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr1_+_6817292 | 0.30 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr9_-_16218097 | 0.30 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr7_+_23495986 | 0.29 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr21_-_7178348 | 0.29 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr7_-_40122139 | 0.29 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr16_+_43152727 | 0.29 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr12_+_11080776 | 0.29 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr17_+_50127648 | 0.29 |
ENSDART00000156460
|
galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr17_+_25289431 | 0.28 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr19_-_41213718 | 0.28 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr20_-_45060241 | 0.28 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr22_+_14051245 | 0.28 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr23_-_35694461 | 0.28 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr25_-_7686201 | 0.28 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr20_-_20932760 | 0.28 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_-_7842078 | 0.27 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr7_-_24373662 | 0.27 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr17_-_33289304 | 0.27 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr10_-_22845485 | 0.27 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr21_-_39628771 | 0.27 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr20_+_25911342 | 0.27 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr11_-_25213651 | 0.26 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr5_+_1278092 | 0.26 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr15_-_33904831 | 0.25 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr3_-_18711288 | 0.25 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr3_+_15279016 | 0.25 |
ENSDART00000182005
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr20_-_45062514 | 0.25 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr14_-_9350364 | 0.24 |
ENSDART00000164451
|
fam46d
|
family with sequence similarity 46, member D |
| chr21_+_18353703 | 0.24 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr22_+_12361317 | 0.24 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr19_-_15397946 | 0.24 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr24_-_14712427 | 0.24 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr11_-_8320868 | 0.24 |
ENSDART00000160570
|
si:ch211-247n2.1
|
si:ch211-247n2.1 |
| chr12_+_9342502 | 0.24 |
ENSDART00000152808
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr3_-_29688370 | 0.23 |
ENSDART00000151147
ENSDART00000151679 |
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr5_-_63109232 | 0.23 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr22_+_12361489 | 0.23 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
| chr5_-_68495967 | 0.23 |
ENSDART00000188107
|
ephb4a
|
eph receptor B4a |
| chr22_+_14051894 | 0.23 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr11_-_41996957 | 0.23 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr11_-_29833698 | 0.23 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr8_+_13368150 | 0.22 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr1_-_44704261 | 0.22 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr17_-_19963718 | 0.22 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr19_-_7406933 | 0.22 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr23_+_34321237 | 0.22 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr15_+_22267847 | 0.22 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr22_-_26236188 | 0.21 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr16_+_5184402 | 0.21 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr6_-_43092175 | 0.21 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr5_+_36870737 | 0.21 |
ENSDART00000145182
|
slc8a2a
|
solute carrier family 8 (sodium/calcium exchanger), member 2a |
| chr17_+_51499789 | 0.21 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr15_+_16521785 | 0.21 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr23_-_10048533 | 0.21 |
ENSDART00000166663
|
plxnb1a
|
plexin b1a |
| chr21_-_12036134 | 0.21 |
ENSDART00000031658
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr16_+_50741154 | 0.21 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr14_-_41285392 | 0.20 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr10_-_3258073 | 0.20 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr20_-_31238313 | 0.20 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr6_+_54498220 | 0.20 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr11_+_15931188 | 0.20 |
ENSDART00000165768
ENSDART00000081098 |
draxin
|
dorsal inhibitory axon guidance protein |
| chr9_+_34641237 | 0.20 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr7_-_34329527 | 0.20 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr3_+_18097700 | 0.20 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr14_-_17563773 | 0.20 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr1_+_53321878 | 0.19 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr16_-_8927425 | 0.19 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr1_-_39977551 | 0.19 |
ENSDART00000139354
|
stox2a
|
storkhead box 2a |
| chr21_-_2248239 | 0.19 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr7_-_18601206 | 0.19 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr2_-_9260002 | 0.19 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr1_-_656693 | 0.19 |
ENSDART00000170483
ENSDART00000166786 |
appa
|
amyloid beta (A4) precursor protein a |
| chr3_+_18437397 | 0.19 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr17_-_6399920 | 0.19 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr14_+_6954579 | 0.19 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr3_-_39190317 | 0.18 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr24_-_25673405 | 0.18 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr1_-_23110740 | 0.18 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr9_-_46276626 | 0.18 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr12_-_30032188 | 0.18 |
ENSDART00000042514
|
atrnl1b
|
attractin-like 1b |
| chr14_+_29609245 | 0.18 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr19_+_42886413 | 0.18 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr20_+_18945057 | 0.18 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr7_+_22688781 | 0.17 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr21_+_21791343 | 0.17 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr6_-_12314475 | 0.17 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr14_-_28534563 | 0.17 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr15_-_30815826 | 0.17 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr2_-_10062575 | 0.17 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr23_+_18779043 | 0.17 |
ENSDART00000158267
|
emp3b
|
epithelial membrane protein 3b |
| chr17_+_24222190 | 0.17 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr18_+_30915348 | 0.17 |
ENSDART00000139398
|
mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr1_-_46981134 | 0.16 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr18_-_16181952 | 0.16 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr7_+_34297271 | 0.16 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_-_31103906 | 0.16 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr1_+_49878000 | 0.16 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr9_-_30346279 | 0.16 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr11_+_6152643 | 0.16 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr2_-_42843797 | 0.16 |
ENSDART00000137913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr10_+_33627429 | 0.16 |
ENSDART00000138542
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr16_-_31598771 | 0.16 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr5_-_33606729 | 0.16 |
ENSDART00000137073
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr6_-_35472923 | 0.16 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr25_+_23358924 | 0.16 |
ENSDART00000156965
|
osbpl5
|
oxysterol binding protein-like 5 |
| chr19_-_25114701 | 0.15 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr18_+_38807239 | 0.15 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr5_+_45990046 | 0.15 |
ENSDART00000084024
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr17_-_15382704 | 0.15 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr6_+_30091811 | 0.15 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr11_-_36341189 | 0.15 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr2_+_51818039 | 0.15 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr8_-_43120844 | 0.15 |
ENSDART00000193242
|
ccdc92
|
coiled-coil domain containing 92 |
| chr5_-_54790923 | 0.15 |
ENSDART00000176835
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_14551077 | 0.15 |
ENSDART00000188717
|
FO704673.1
|
|
| chr24_-_12832499 | 0.15 |
ENSDART00000182027
ENSDART00000039312 |
ipo4
|
importin 4 |
| chr17_-_43552894 | 0.15 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr3_-_40976288 | 0.14 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr17_+_51520073 | 0.14 |
ENSDART00000189646
ENSDART00000170951 ENSDART00000189492 |
pxdn
|
peroxidasin |
| chr13_+_41917606 | 0.14 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr1_-_54938137 | 0.14 |
ENSDART00000147212
|
crtac1a
|
cartilage acidic protein 1a |
| chr14_+_25817628 | 0.14 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr25_+_20140926 | 0.14 |
ENSDART00000121989
|
cald1b
|
caldesmon 1b |
| chr5_-_5243079 | 0.14 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr15_+_28368823 | 0.14 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr9_+_31534601 | 0.14 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr11_-_472547 | 0.14 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr1_-_49649122 | 0.14 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr3_+_12334509 | 0.14 |
ENSDART00000165796
|
glis2b
|
GLIS family zinc finger 2b |
| chr22_-_26175237 | 0.14 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr23_-_16960972 | 0.14 |
ENSDART00000142546
|
cdc42l2
|
cell division cycle 42 like 2 |
| chr25_+_26844028 | 0.14 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr4_+_9030609 | 0.13 |
ENSDART00000154399
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr7_+_65876335 | 0.13 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr11_+_25259058 | 0.13 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_-_36234185 | 0.13 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr12_-_41662327 | 0.13 |
ENSDART00000191602
ENSDART00000170423 |
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr11_-_27537593 | 0.13 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr25_+_25123385 | 0.13 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr3_+_56366395 | 0.13 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr7_-_35314347 | 0.13 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr14_+_3411771 | 0.13 |
ENSDART00000164778
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr15_+_28368644 | 0.13 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr19_+_32947910 | 0.12 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase H+ transporting V1 subunit C1b |
| chr4_-_42074960 | 0.12 |
ENSDART00000164406
|
zgc:174696
|
zgc:174696 |
| chr11_-_36341028 | 0.12 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr14_-_13048355 | 0.12 |
ENSDART00000166434
|
si:dkey-35h6.1
|
si:dkey-35h6.1 |
| chr12_-_16452200 | 0.12 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr14_+_11909966 | 0.12 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr1_+_29281764 | 0.12 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr21_-_37972412 | 0.12 |
ENSDART00000186315
|
ripply1
|
ripply transcriptional repressor 1 |
| chr18_+_17534627 | 0.12 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr6_-_44402358 | 0.12 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr23_+_42346799 | 0.12 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr16_-_30826712 | 0.12 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr14_-_46113321 | 0.12 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr8_+_24747865 | 0.12 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr25_+_19201231 | 0.12 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr13_+_2617555 | 0.12 |
ENSDART00000162208
|
plpp4
|
phospholipid phosphatase 4 |
| chr21_-_39546737 | 0.11 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr19_+_42432625 | 0.11 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr15_+_1765988 | 0.11 |
ENSDART00000159032
|
cul3b
|
cullin 3b |
| chr25_-_21280584 | 0.11 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr13_-_10730056 | 0.11 |
ENSDART00000126725
|
ppm1ba
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ba |
| chr3_-_40976463 | 0.11 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr14_-_30897177 | 0.11 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr14_+_36246726 | 0.11 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr24_+_26997798 | 0.11 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr21_-_20381481 | 0.11 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr25_-_13490744 | 0.11 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr11_-_45385602 | 0.11 |
ENSDART00000166691
|
trappc10
|
trafficking protein particle complex 10 |
| chr13_+_40034176 | 0.11 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr21_+_41511469 | 0.11 |
ENSDART00000172563
|
stc2b
|
stanniocalcin 2b |
| chr17_-_5769196 | 0.11 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr11_-_1948784 | 0.11 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr5_-_37871526 | 0.11 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr16_-_27138478 | 0.11 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr21_+_39963851 | 0.10 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr10_-_34981219 | 0.10 |
ENSDART00000132823
|
smad9
|
SMAD family member 9 |
| chr16_-_52736549 | 0.10 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.2 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.1 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0019068 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.2 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0071294 | cellular response to cadmium ion(GO:0071276) cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |