Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
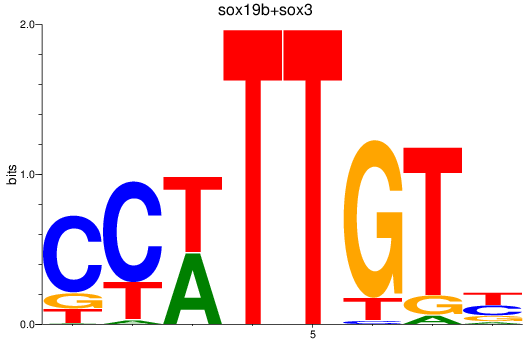
Results for sox19b+sox3
Z-value: 0.54
Transcription factors associated with sox19b+sox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19b
|
ENSDARG00000040266 | SRY-box transcription factor 19b |
|
sox3
|
ENSDARG00000053569 | SRY-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox19b | dr11_v1_chr7_-_26497947_26497947 | -0.93 | 2.0e-02 | Click! |
| sox3 | dr11_v1_chr14_-_32744464_32744464 | -0.78 | 1.2e-01 | Click! |
Activity profile of sox19b+sox3 motif
Sorted Z-values of sox19b+sox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_14280598 | 0.42 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr2_-_24269911 | 0.38 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr23_+_20110086 | 0.36 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr2_-_24270062 | 0.36 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr9_-_23891102 | 0.35 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr24_-_40668208 | 0.34 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr5_-_46505691 | 0.34 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr19_+_46113828 | 0.34 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr20_-_33976305 | 0.34 |
ENSDART00000111022
|
sele
|
selectin E |
| chr10_+_33171501 | 0.33 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr23_-_10175898 | 0.30 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr11_+_10548171 | 0.29 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr23_+_5499307 | 0.29 |
ENSDART00000170361
|
tead3a
|
TEA domain family member 3 a |
| chr3_+_3545825 | 0.29 |
ENSDART00000109060
|
CR589947.1
|
|
| chr6_+_54576520 | 0.28 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr9_+_23770666 | 0.27 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr12_-_30777743 | 0.27 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr25_+_35502552 | 0.27 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr20_+_40237441 | 0.27 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr16_+_29586468 | 0.26 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr16_-_33104944 | 0.26 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr15_-_43164591 | 0.26 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr22_-_10541372 | 0.26 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr8_-_18537866 | 0.26 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr25_+_14109626 | 0.26 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr23_-_31506854 | 0.26 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr22_+_19220459 | 0.25 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr14_+_11457500 | 0.25 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr19_+_20778011 | 0.25 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr7_+_35268880 | 0.25 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr12_-_36045283 | 0.25 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr19_+_24488403 | 0.25 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr15_-_37829160 | 0.25 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr25_-_22187397 | 0.24 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr7_-_53117131 | 0.24 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr13_+_30225262 | 0.24 |
ENSDART00000141945
|
rps24
|
ribosomal protein S24 |
| chr7_-_6459481 | 0.23 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr4_+_18843015 | 0.23 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr17_+_25849332 | 0.23 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr9_-_21912227 | 0.23 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr3_-_34069637 | 0.23 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr23_-_5683147 | 0.22 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr10_+_9550419 | 0.22 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr3_-_57425961 | 0.22 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr13_+_22675802 | 0.22 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr16_-_32975951 | 0.22 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr2_-_37103622 | 0.22 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr18_-_6633984 | 0.22 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr6_+_52804267 | 0.21 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr18_+_8912113 | 0.21 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr18_-_6634424 | 0.21 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr6_+_45932276 | 0.21 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_-_23256726 | 0.21 |
ENSDART00000131353
|
si:dkey-98j1.5
|
si:dkey-98j1.5 |
| chr21_+_19368720 | 0.21 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr7_+_48761646 | 0.20 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr7_+_39410180 | 0.20 |
ENSDART00000168641
|
CT030188.1
|
|
| chr16_-_31598771 | 0.20 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr5_-_7834582 | 0.20 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr11_-_10659195 | 0.20 |
ENSDART00000115255
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr14_-_8080416 | 0.20 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr16_+_35535375 | 0.20 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr9_-_50001606 | 0.20 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr7_+_48761875 | 0.20 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr22_+_661711 | 0.19 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr20_+_23498255 | 0.19 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr21_+_11776328 | 0.19 |
ENSDART00000134469
ENSDART00000081646 |
glrx
|
glutaredoxin (thioltransferase) |
| chr12_-_30777540 | 0.19 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr18_+_8912710 | 0.19 |
ENSDART00000142866
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr23_+_36144487 | 0.19 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr5_-_37117778 | 0.19 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_+_22273123 | 0.19 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr9_+_13986427 | 0.19 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr4_-_16354292 | 0.19 |
ENSDART00000139919
|
lum
|
lumican |
| chr16_+_25245857 | 0.18 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr21_-_11646878 | 0.18 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr3_-_50139860 | 0.18 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr8_-_43750062 | 0.18 |
ENSDART00000142243
|
ulk1a
|
unc-51 like autophagy activating kinase 1a |
| chr11_-_28050559 | 0.18 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr19_+_47394270 | 0.18 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr19_+_43119698 | 0.18 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr3_+_29640996 | 0.18 |
ENSDART00000011052
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr14_+_38786298 | 0.17 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr7_+_39410393 | 0.17 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr3_+_8290595 | 0.17 |
ENSDART00000142691
|
trim35-10
|
tripartite motif containing 35-10 |
| chr15_-_3736149 | 0.17 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr7_+_39444843 | 0.17 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr13_+_46941930 | 0.17 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr6_-_13206255 | 0.17 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr12_-_22524388 | 0.17 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr17_+_45405821 | 0.17 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr23_-_30045661 | 0.17 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr23_-_21515182 | 0.17 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr5_+_15203421 | 0.17 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr18_+_8912536 | 0.16 |
ENSDART00000134827
ENSDART00000061904 |
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr5_+_4564233 | 0.16 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr5_-_55560937 | 0.16 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr2_-_51794472 | 0.16 |
ENSDART00000186652
|
BX908782.3
|
|
| chr23_-_33738945 | 0.16 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr23_-_24263474 | 0.16 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr24_+_20373605 | 0.16 |
ENSDART00000112349
|
plcd1a
|
phospholipase C, delta 1a |
| chr15_-_4528326 | 0.16 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr3_-_45326897 | 0.16 |
ENSDART00000086417
|
il21r.2
|
interleukin 21 receptor, tandem duplicate 2 |
| chr16_+_21801277 | 0.16 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr9_-_43082945 | 0.16 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr15_+_37331585 | 0.16 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr17_+_30546579 | 0.16 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr21_-_30166097 | 0.16 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr15_-_36533322 | 0.16 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr14_-_34059681 | 0.15 |
ENSDART00000003993
|
itk
|
IL2 inducible T cell kinase |
| chr23_+_42813415 | 0.15 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr8_-_28449782 | 0.15 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr6_-_10233538 | 0.15 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr19_-_19721556 | 0.15 |
ENSDART00000165196
|
evx1
|
even-skipped homeobox 1 |
| chr12_-_29305533 | 0.15 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr14_+_46313135 | 0.15 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr23_-_3758637 | 0.15 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr1_-_33645967 | 0.15 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr6_-_52675630 | 0.15 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr16_+_26732086 | 0.15 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr24_+_26932472 | 0.15 |
ENSDART00000192288
ENSDART00000079848 |
tnfsf10
|
TNF superfamily member 10 |
| chr3_+_12755535 | 0.15 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr3_+_57820913 | 0.15 |
ENSDART00000168101
|
CU571328.1
|
|
| chr16_+_35535171 | 0.15 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr18_-_38270430 | 0.15 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr16_-_41762983 | 0.15 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr22_-_10121880 | 0.15 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_-_21810668 | 0.15 |
ENSDART00000156575
|
dicp3.3
|
diverse immunoglobulin domain-containing protein 3.3 |
| chr1_-_25936677 | 0.15 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr9_-_14084743 | 0.14 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr4_-_2380173 | 0.14 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr18_+_35842933 | 0.14 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr10_-_25816558 | 0.14 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr25_+_31323978 | 0.14 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr1_-_58111826 | 0.14 |
ENSDART00000134949
|
caspbl
|
caspase b, like |
| chr5_+_25762271 | 0.14 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr25_-_27843066 | 0.14 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr7_+_38380135 | 0.14 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr3_-_59297532 | 0.14 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr6_-_43677125 | 0.14 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr12_-_30523216 | 0.14 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr10_+_8690936 | 0.14 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr24_-_11076400 | 0.14 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr7_-_8408014 | 0.14 |
ENSDART00000112492
|
zgc:194686
|
zgc:194686 |
| chr5_-_30984010 | 0.14 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr25_-_35124576 | 0.14 |
ENSDART00000128017
|
zgc:171759
|
zgc:171759 |
| chr8_+_14158021 | 0.14 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr8_-_23884301 | 0.14 |
ENSDART00000185509
ENSDART00000147202 |
lhfpl5b
|
LHFPL tetraspan subfamily member 5b |
| chr7_-_7398350 | 0.14 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr25_-_3470910 | 0.14 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr14_+_30910114 | 0.14 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr11_+_30295582 | 0.14 |
ENSDART00000122424
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr2_-_26596794 | 0.14 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr5_-_30615901 | 0.14 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr25_-_12998202 | 0.14 |
ENSDART00000171661
|
ccl39.3
|
chemokine (C-C motif) ligand 39, duplicate 3 |
| chr24_-_31194847 | 0.14 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr6_-_9707599 | 0.14 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr3_+_3573696 | 0.14 |
ENSDART00000169680
|
CR589947.2
|
|
| chr3_-_32956808 | 0.14 |
ENSDART00000183902
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr1_-_47431453 | 0.14 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr11_+_24716837 | 0.14 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr21_+_27382893 | 0.14 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr23_+_24085531 | 0.14 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr7_-_28681724 | 0.13 |
ENSDART00000162400
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr5_-_41841892 | 0.13 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr15_-_29586747 | 0.13 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr16_-_26731928 | 0.13 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr20_+_13533544 | 0.13 |
ENSDART00000143115
|
sytl3
|
synaptotagmin-like 3 |
| chr2_-_19234329 | 0.13 |
ENSDART00000161106
ENSDART00000160060 ENSDART00000174552 |
cdc20
|
cell division cycle 20 homolog |
| chr4_-_11403811 | 0.13 |
ENSDART00000067272
ENSDART00000140018 |
pimr173
|
Pim proto-oncogene, serine/threonine kinase, related 173 |
| chr9_-_22831836 | 0.13 |
ENSDART00000142585
|
neb
|
nebulin |
| chr7_+_48555400 | 0.13 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr8_-_28349859 | 0.13 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr1_+_22654875 | 0.13 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr18_-_38270077 | 0.13 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr6_-_13498745 | 0.13 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr5_+_38680247 | 0.13 |
ENSDART00000132763
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr23_+_42819221 | 0.13 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr9_-_24244383 | 0.13 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr3_-_31258459 | 0.13 |
ENSDART00000177928
|
LO017965.1
|
|
| chr23_-_3759345 | 0.13 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr18_+_44649804 | 0.13 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr1_-_59124505 | 0.13 |
ENSDART00000186257
|
FO203432.1
|
|
| chr24_-_2947393 | 0.13 |
ENSDART00000166661
ENSDART00000147110 |
tubb6
|
tubulin, beta 6 class V |
| chr25_-_3830272 | 0.13 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr11_+_29965822 | 0.13 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr23_-_19715799 | 0.13 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr16_-_31976269 | 0.13 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr15_+_12429206 | 0.13 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr25_-_35145920 | 0.13 |
ENSDART00000157137
|
si:dkey-261m9.19
|
si:dkey-261m9.19 |
| chr2_+_30721070 | 0.13 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr24_+_26006730 | 0.13 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr9_+_50001746 | 0.13 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr25_-_35106751 | 0.13 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr16_+_35594670 | 0.13 |
ENSDART00000163275
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr19_+_10603405 | 0.13 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr11_-_6048490 | 0.13 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr8_+_48491387 | 0.12 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr19_+_4051695 | 0.12 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr16_-_27566552 | 0.12 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr13_-_17723417 | 0.12 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19b+sox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.5 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0043282 | pharyngeal muscle development(GO:0043282) |
| 0.0 | 0.2 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.0 | 0.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.0 | 0.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0090190 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.1 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.0 | 0.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0034473 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.0 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.0 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |