Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
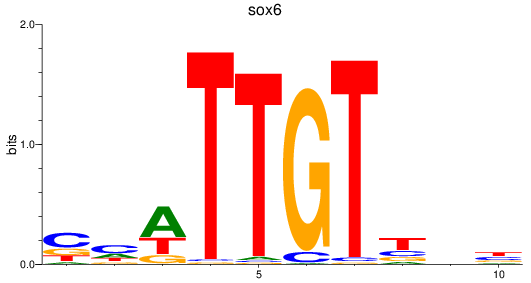
Results for sox6
Z-value: 0.68
Transcription factors associated with sox6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox6
|
ENSDARG00000015536 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox6 | dr11_v1_chr7_+_27251376_27251475 | -0.12 | 8.5e-01 | Click! |
Activity profile of sox6 motif
Sorted Z-values of sox6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_48340133 | 0.62 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr10_+_15608326 | 0.51 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr1_-_55810730 | 0.41 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr5_+_35458190 | 0.38 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr24_-_40668208 | 0.35 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr15_+_28368823 | 0.34 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr16_+_25245857 | 0.34 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr20_-_40754794 | 0.31 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr22_-_24967348 | 0.31 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr13_-_20540790 | 0.31 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr7_+_39444843 | 0.30 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr16_+_47308856 | 0.26 |
ENSDART00000170103
|
col28a1b
|
collagen, type XXVIII, alpha 1b |
| chr20_+_40237441 | 0.25 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr2_+_16780643 | 0.25 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr24_-_29868151 | 0.25 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr8_+_17069577 | 0.24 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr12_-_22524388 | 0.23 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr5_+_10046643 | 0.23 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr9_-_23891102 | 0.23 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr5_-_46505691 | 0.23 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr25_+_26798673 | 0.22 |
ENSDART00000157235
|
ca12
|
carbonic anhydrase XII |
| chr15_-_30853246 | 0.22 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr21_-_35853245 | 0.22 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr19_-_23249822 | 0.22 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr14_+_51056605 | 0.21 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr22_+_1028724 | 0.21 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr12_-_33354409 | 0.19 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr4_-_77432218 | 0.19 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr14_+_48812189 | 0.18 |
ENSDART00000161454
|
mmp17b
|
matrix metallopeptidase 17b |
| chr13_+_33368503 | 0.18 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr3_+_29641181 | 0.18 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr5_+_35458346 | 0.16 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr17_-_2578026 | 0.15 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_-_33961697 | 0.15 |
ENSDART00000061765
|
selp
|
selectin P |
| chr17_-_2573021 | 0.15 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr8_+_14158021 | 0.14 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr7_-_17570923 | 0.14 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr1_-_354115 | 0.13 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr2_-_24269911 | 0.13 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr10_-_22831611 | 0.13 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr1_-_17735861 | 0.13 |
ENSDART00000018238
|
pdlim3a
|
PDZ and LIM domain 3a |
| chr16_-_21799407 | 0.13 |
ENSDART00000123717
|
FP015862.4
|
|
| chr23_-_25798099 | 0.13 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr15_-_35410860 | 0.12 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr19_-_10771558 | 0.12 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr13_-_7567707 | 0.12 |
ENSDART00000190296
ENSDART00000180348 |
pitx3
|
paired-like homeodomain 3 |
| chr25_+_13620555 | 0.12 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr14_-_41478265 | 0.12 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr5_+_55221593 | 0.11 |
ENSDART00000073638
|
tmc2a
|
transmembrane channel-like 2a |
| chr25_+_35502552 | 0.11 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr5_-_33606729 | 0.11 |
ENSDART00000137073
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr20_+_37295006 | 0.11 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr2_-_24270062 | 0.11 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr11_-_9808356 | 0.11 |
ENSDART00000167465
|
nlgn1
|
neuroligin 1 |
| chr21_-_41870029 | 0.11 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr3_-_16250527 | 0.10 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr24_-_19719240 | 0.10 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr7_+_38255418 | 0.10 |
ENSDART00000052354
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr15_-_18067220 | 0.10 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr6_-_52675630 | 0.10 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr3_+_49074008 | 0.10 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr22_-_10440688 | 0.10 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr23_+_3538463 | 0.10 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr22_-_37565348 | 0.10 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr5_+_7989210 | 0.10 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr23_+_27068225 | 0.09 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr11_+_31324335 | 0.09 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr17_-_2590222 | 0.09 |
ENSDART00000185711
|
CR759892.1
|
|
| chr8_+_42941555 | 0.09 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr3_+_3099492 | 0.09 |
ENSDART00000154156
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr19_+_24575077 | 0.09 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr20_-_13722514 | 0.09 |
ENSDART00000152458
|
si:ch211-122h15.4
|
si:ch211-122h15.4 |
| chr18_+_21122818 | 0.09 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr4_+_18804317 | 0.09 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr10_-_26163989 | 0.09 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr7_+_48761875 | 0.08 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr1_-_33645967 | 0.08 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr14_-_34771371 | 0.08 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr5_-_66160415 | 0.08 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr9_-_22272181 | 0.08 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr3_-_21402279 | 0.08 |
ENSDART00000164513
|
CT573446.1
|
|
| chr22_+_3045495 | 0.08 |
ENSDART00000164061
|
LO017843.1
|
|
| chr2_+_23731194 | 0.08 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr14_-_33978117 | 0.08 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr12_+_19320657 | 0.08 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr9_+_50001746 | 0.08 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr5_+_19097194 | 0.08 |
ENSDART00000131918
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr7_+_48761646 | 0.07 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr4_+_65607540 | 0.07 |
ENSDART00000192218
|
si:dkey-205i10.2
|
si:dkey-205i10.2 |
| chr10_+_39200213 | 0.07 |
ENSDART00000153727
|
ei24
|
etoposide induced 2.4 |
| chr8_+_17869225 | 0.07 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr7_+_22688781 | 0.07 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr1_+_17376922 | 0.07 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr11_+_10541258 | 0.07 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr5_-_20195350 | 0.06 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr14_+_52565660 | 0.06 |
ENSDART00000188151
|
rpl26
|
ribosomal protein L26 |
| chr16_-_42186093 | 0.06 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr23_+_5499307 | 0.06 |
ENSDART00000170361
|
tead3a
|
TEA domain family member 3 a |
| chr22_-_12746539 | 0.06 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr16_+_53387085 | 0.05 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr5_-_31875645 | 0.05 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr25_+_28567381 | 0.05 |
ENSDART00000126552
|
SLC15A5
|
si:ch211-190o6.3 |
| chr25_+_13271458 | 0.05 |
ENSDART00000188307
|
AL772241.1
|
|
| chr10_+_16069987 | 0.05 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr2_-_5942115 | 0.05 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr4_-_76154252 | 0.05 |
ENSDART00000161269
|
BX957252.1
|
|
| chr17_-_45386546 | 0.05 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr23_-_21471022 | 0.05 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_14084044 | 0.05 |
ENSDART00000141571
|
fer1l6
|
fer-1-like family member 6 |
| chr19_-_27339844 | 0.05 |
ENSDART00000052358
ENSDART00000148238 ENSDART00000147661 ENSDART00000137346 |
znrd1
|
zinc ribbon domain containing 1 |
| chr1_-_50791280 | 0.05 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr6_-_41138854 | 0.05 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr4_+_29702703 | 0.05 |
ENSDART00000167771
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr21_+_36396864 | 0.04 |
ENSDART00000137309
|
gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr11_-_7147540 | 0.04 |
ENSDART00000143942
|
bmp7a
|
bone morphogenetic protein 7a |
| chr19_-_27588842 | 0.04 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr21_+_34119759 | 0.04 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr25_-_16076257 | 0.04 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr23_-_21453614 | 0.04 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr12_+_42400277 | 0.04 |
ENSDART00000166590
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr2_-_10896745 | 0.04 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr23_-_21463788 | 0.04 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr13_-_11699530 | 0.04 |
ENSDART00000192161
|
slc39a8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr7_+_65876335 | 0.04 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr1_-_31534089 | 0.04 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr16_+_27349585 | 0.04 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr15_-_20779624 | 0.04 |
ENSDART00000181936
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr3_+_29640996 | 0.04 |
ENSDART00000011052
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr17_-_20228610 | 0.04 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr11_-_3381343 | 0.04 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr17_-_45386823 | 0.04 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr13_+_42309688 | 0.04 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr12_+_38697541 | 0.03 |
ENSDART00000188648
|
dnai2b
|
dynein, axonemal, intermediate chain 2b |
| chr16_-_22006996 | 0.03 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr7_-_6459481 | 0.03 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr4_-_63848305 | 0.03 |
ENSDART00000097308
|
BX901974.1
|
|
| chr23_-_7826849 | 0.03 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr21_-_2310064 | 0.03 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr15_-_5563551 | 0.03 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr10_+_44042033 | 0.03 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr2_+_26479676 | 0.03 |
ENSDART00000056795
ENSDART00000144837 |
hectd3
|
HECT domain containing 3 |
| chr10_+_5744941 | 0.03 |
ENSDART00000159769
ENSDART00000184734 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr8_-_49499457 | 0.03 |
ENSDART00000098326
|
opn7d
|
opsin 7, group member d |
| chr24_+_18714212 | 0.03 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr23_-_5685023 | 0.03 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr15_+_29727799 | 0.03 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr7_+_25103965 | 0.03 |
ENSDART00000192561
ENSDART00000173721 |
si:dkey-23i12.9
|
si:dkey-23i12.9 |
| chr25_-_22889519 | 0.02 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr23_+_21459263 | 0.02 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr25_+_25737386 | 0.02 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr20_+_35445462 | 0.02 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr22_-_18546241 | 0.02 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr15_-_41503392 | 0.02 |
ENSDART00000169351
|
si:dkey-230i18.2
|
si:dkey-230i18.2 |
| chr3_+_3545825 | 0.02 |
ENSDART00000109060
|
CR589947.1
|
|
| chr6_+_54358529 | 0.02 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr23_+_30827959 | 0.02 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr19_+_19762183 | 0.02 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr7_-_24520866 | 0.02 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr2_+_47623202 | 0.02 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr7_+_20019125 | 0.02 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr15_+_19458982 | 0.02 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr14_-_1990290 | 0.02 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr21_+_11105007 | 0.01 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr5_+_40835601 | 0.01 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr19_-_18152407 | 0.01 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr1_-_47431453 | 0.01 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr13_+_46941930 | 0.01 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_+_21722892 | 0.01 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr7_-_24373662 | 0.01 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr3_+_3992590 | 0.01 |
ENSDART00000180211
ENSDART00000054840 |
BX005421.2
|
|
| chr7_-_32895668 | 0.01 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr6_+_58280936 | 0.01 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr7_-_73852594 | 0.01 |
ENSDART00000183194
|
zgc:165555
|
zgc:165555 |
| chr25_-_3503164 | 0.00 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr5_-_38384289 | 0.00 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr12_+_5081759 | 0.00 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr17_+_7534365 | 0.00 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr3_-_26524934 | 0.00 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr10_+_3428194 | 0.00 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr10_+_21677058 | 0.00 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr17_+_26208630 | 0.00 |
ENSDART00000087084
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr7_-_24995631 | 0.00 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr12_+_40810418 | 0.00 |
ENSDART00000183393
|
CDH18
|
cadherin 18 |
| chr23_-_33944597 | 0.00 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr15_+_12429206 | 0.00 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0090189 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.2 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |