Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
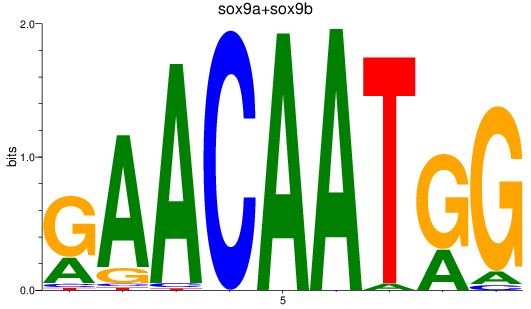
Results for sox9a+sox9b
Z-value: 0.96
Transcription factors associated with sox9a+sox9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox9a
|
ENSDARG00000003293 | SRY-box transcription factor 9a |
|
sox9b
|
ENSDARG00000043923 | SRY-box transcription factor 9b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox9a | dr11_v1_chr12_-_1951233_1951284 | 0.89 | 4.4e-02 | Click! |
| sox9b | dr11_v1_chr3_-_62527675_62527696 | 0.14 | 8.2e-01 | Click! |
Activity profile of sox9a+sox9b motif
Sorted Z-values of sox9a+sox9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 1.70 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr21_-_29100110 | 0.73 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr23_+_35708730 | 0.72 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr17_-_46457622 | 0.72 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr5_-_29488245 | 0.71 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr10_-_31782616 | 0.69 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr23_-_3681026 | 0.67 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr14_-_33872092 | 0.67 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr2_+_3472832 | 0.64 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr2_+_34967210 | 0.62 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr11_+_25735478 | 0.62 |
ENSDART00000103566
|
si:dkey-183j2.10
|
si:dkey-183j2.10 |
| chr6_+_12853655 | 0.62 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr3_-_22212764 | 0.61 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr25_+_8063455 | 0.59 |
ENSDART00000073919
|
kcnc1b
|
potassium voltage-gated channel, Shaw-related subfamily, member 1b |
| chr6_-_43092175 | 0.57 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr4_-_211714 | 0.56 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_31110817 | 0.54 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr11_+_35364445 | 0.54 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr19_+_342094 | 0.52 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr13_+_35746440 | 0.51 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr19_-_103289 | 0.50 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr9_-_31278048 | 0.49 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr16_-_46097432 | 0.49 |
ENSDART00000160071
|
si:ch211-271d10.2
|
si:ch211-271d10.2 |
| chr14_-_1990290 | 0.49 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr19_-_3167729 | 0.49 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr2_+_34967022 | 0.49 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr24_-_21674950 | 0.48 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr7_-_38698583 | 0.47 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr16_+_29303971 | 0.46 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr9_+_17348745 | 0.45 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr2_-_22286828 | 0.45 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr12_+_33151246 | 0.44 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr20_-_40451115 | 0.44 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr20_-_23226453 | 0.44 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr15_-_33896159 | 0.43 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr17_-_14726824 | 0.43 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr14_-_31893996 | 0.43 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr16_+_34523515 | 0.42 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr2_+_26303627 | 0.42 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr17_-_36936649 | 0.42 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr2_+_56463167 | 0.41 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr5_+_36768674 | 0.41 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr23_-_21463788 | 0.40 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr7_-_28147838 | 0.40 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr19_+_30633453 | 0.40 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr2_+_31957554 | 0.40 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr23_-_33654889 | 0.39 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr10_+_22003750 | 0.39 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr5_-_22082918 | 0.39 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr7_+_60551133 | 0.39 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr17_-_36936856 | 0.39 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr21_+_3093419 | 0.38 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr3_+_24134418 | 0.38 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr13_-_638485 | 0.38 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr1_+_18965750 | 0.38 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr4_+_14660769 | 0.38 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_-_74367912 | 0.37 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr8_-_23081511 | 0.36 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr10_-_22249444 | 0.36 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr11_-_27057572 | 0.36 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr7_-_58098814 | 0.36 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr1_-_44701313 | 0.36 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr21_-_43550120 | 0.36 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr17_-_26911852 | 0.36 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr23_-_637347 | 0.35 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr10_+_22381802 | 0.35 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr21_-_11054876 | 0.35 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr3_-_59981162 | 0.34 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr9_-_18742704 | 0.34 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr16_-_19890303 | 0.34 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr6_-_12172424 | 0.34 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr16_+_37582872 | 0.33 |
ENSDART00000169331
|
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr1_-_40994259 | 0.33 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr15_-_33925851 | 0.33 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr10_+_18952271 | 0.33 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr12_-_31009315 | 0.33 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr10_-_26163989 | 0.33 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr6_-_51101834 | 0.33 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr21_-_26089964 | 0.33 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr24_-_22756508 | 0.33 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr18_+_45571378 | 0.32 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr10_+_15255012 | 0.32 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr3_+_45401472 | 0.32 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr16_+_5678071 | 0.32 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr6_+_29410986 | 0.32 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr5_-_41560874 | 0.31 |
ENSDART00000136702
|
dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr16_-_6821927 | 0.31 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr23_+_19590006 | 0.31 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr19_-_3488860 | 0.31 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr3_-_50277959 | 0.30 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr6_-_12851888 | 0.30 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr8_+_23093155 | 0.30 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr21_+_32820175 | 0.30 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr10_+_15255198 | 0.30 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr25_-_29134654 | 0.30 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr15_-_35246742 | 0.30 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr2_-_22230326 | 0.30 |
ENSDART00000127810
|
fam110b
|
family with sequence similarity 110, member B |
| chr24_-_7699356 | 0.30 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr9_+_6009077 | 0.30 |
ENSDART00000057484
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr17_-_29119362 | 0.30 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr3_+_54047342 | 0.29 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr18_+_38900092 | 0.29 |
ENSDART00000148541
ENSDART00000133618 ENSDART00000088037 |
myo5aa
|
myosin VAa |
| chr24_-_19719240 | 0.29 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr5_+_24156170 | 0.29 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr6_-_37622576 | 0.29 |
ENSDART00000154363
|
chst7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr15_-_16679517 | 0.29 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr7_-_33023404 | 0.29 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr1_-_46832880 | 0.28 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr4_+_74551606 | 0.28 |
ENSDART00000174339
|
kcna1b
|
potassium voltage-gated channel, shaker-related subfamily, member 1b |
| chr23_+_28648864 | 0.28 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr7_-_51627019 | 0.28 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr4_-_9586713 | 0.28 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr7_-_28148310 | 0.28 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr7_-_51476276 | 0.28 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr23_-_21453614 | 0.28 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr18_-_16181952 | 0.27 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr6_-_21873266 | 0.27 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr9_+_49659114 | 0.27 |
ENSDART00000189643
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr5_-_23999777 | 0.27 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr17_-_23241393 | 0.27 |
ENSDART00000190697
|
AL935174.4
|
|
| chr13_+_12739283 | 0.27 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr13_+_28732101 | 0.27 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr21_+_17301790 | 0.27 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr22_+_5106751 | 0.27 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr17_-_37054959 | 0.26 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr23_-_14918276 | 0.26 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr23_-_32892441 | 0.26 |
ENSDART00000147998
|
plxna2
|
plexin A2 |
| chr20_+_54383838 | 0.26 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr7_-_69636502 | 0.26 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr3_-_42086577 | 0.26 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr8_+_26879358 | 0.26 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr19_-_34011340 | 0.26 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr24_+_27023274 | 0.26 |
ENSDART00000137212
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr3_+_24189804 | 0.26 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr5_-_11943750 | 0.26 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr16_+_34522977 | 0.25 |
ENSDART00000144069
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr8_+_21146262 | 0.25 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr21_+_25057176 | 0.25 |
ENSDART00000183975
|
zgc:171740
|
zgc:171740 |
| chr9_-_7212973 | 0.25 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr25_+_37126921 | 0.25 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr5_-_48260145 | 0.25 |
ENSDART00000044083
ENSDART00000163250 ENSDART00000135911 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr19_+_47311020 | 0.25 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr5_-_6377865 | 0.25 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr11_+_6152643 | 0.25 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr4_-_837768 | 0.25 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr19_+_47311869 | 0.25 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr17_-_42097267 | 0.24 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr6_+_54358529 | 0.24 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr3_+_22591395 | 0.24 |
ENSDART00000190526
|
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr19_-_43226124 | 0.24 |
ENSDART00000168965
|
gnrhr1
|
gonadotropin releasing hormone receptor 1 |
| chr18_-_1185772 | 0.24 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr16_-_22781446 | 0.24 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr2_-_33645411 | 0.24 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr24_-_35699595 | 0.24 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_-_20511595 | 0.24 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr17_-_3190070 | 0.24 |
ENSDART00000115027
|
tmem151bb
|
transmembrane protein 151Bb |
| chr8_+_25267903 | 0.24 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr17_+_43486436 | 0.24 |
ENSDART00000023953
ENSDART00000149041 |
reep1
|
receptor accessory protein 1 |
| chr7_-_6604623 | 0.24 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr5_+_59397739 | 0.23 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr17_+_44780166 | 0.23 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr3_+_29600917 | 0.23 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr11_+_24925434 | 0.23 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr7_+_22801465 | 0.23 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr4_-_11737939 | 0.23 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr19_+_7549854 | 0.23 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr8_-_31393429 | 0.23 |
ENSDART00000192369
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr6_+_2951645 | 0.23 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr12_+_24561832 | 0.23 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr20_-_31238313 | 0.22 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr14_-_33095917 | 0.22 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr7_-_18601206 | 0.22 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr14_-_2217285 | 0.22 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr3_-_21062706 | 0.22 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr1_-_46981134 | 0.22 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_-_30787932 | 0.22 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr25_-_13789955 | 0.21 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr5_-_60159116 | 0.21 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr9_+_34641237 | 0.21 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr3_+_31396149 | 0.21 |
ENSDART00000151423
ENSDART00000193580 |
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr1_+_49878000 | 0.21 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr18_+_31280984 | 0.21 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr21_+_21791343 | 0.21 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr3_+_40856095 | 0.21 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr11_+_13067028 | 0.21 |
ENSDART00000186126
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr21_-_5958387 | 0.21 |
ENSDART00000130521
|
CACFD1
|
si:ch73-209e20.5 |
| chr10_-_25628555 | 0.21 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr23_-_33944597 | 0.21 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr8_+_25254435 | 0.21 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr22_-_22416337 | 0.21 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr10_+_2568793 | 0.21 |
ENSDART00000191985
ENSDART00000081922 |
spina
|
spindlin a |
| chr20_-_6184167 | 0.21 |
ENSDART00000147451
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr21_-_43485351 | 0.21 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr14_-_44841335 | 0.21 |
ENSDART00000173011
|
GRXCR1
|
si:dkey-109l4.6 |
| chr14_+_23717165 | 0.21 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_-_2318590 | 0.21 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr2_+_24868010 | 0.21 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr24_+_37598441 | 0.21 |
ENSDART00000125145
|
rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr22_+_15507218 | 0.20 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr9_+_8942258 | 0.20 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox9a+sox9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.6 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.5 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.8 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.3 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.3 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.2 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.5 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 0.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.0 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.8 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.0 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.0 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of sulfur metabolic process(GO:0042762) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.0 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.4 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.3 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |